HIV-1gp120 gene consensus sequence optimized by codon and gp120 nucleic acid vaccine
A HIV-1, codon optimization technology, applied in gene therapy, genetic engineering, plant gene improvement, etc., to achieve a good immune response
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
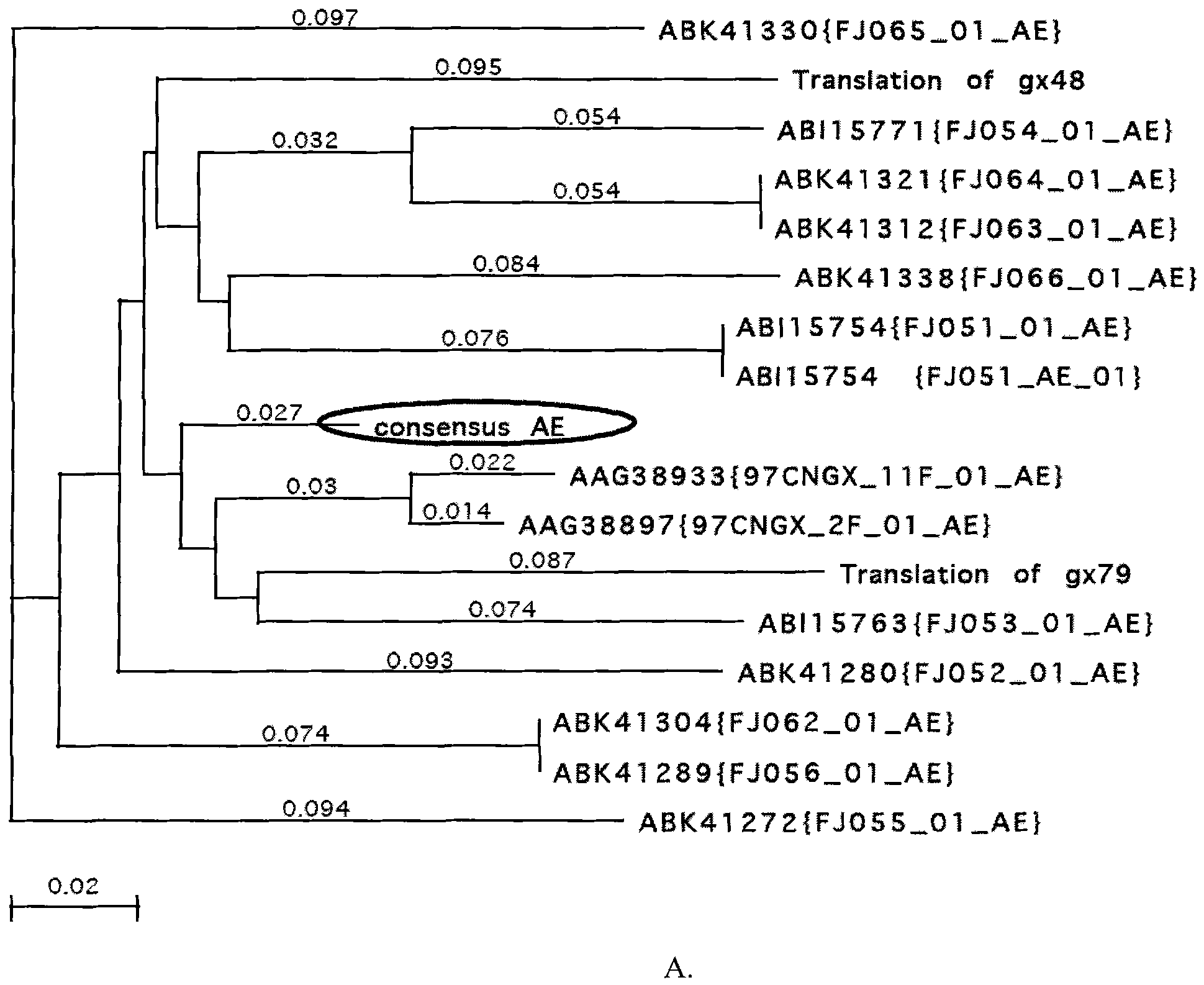
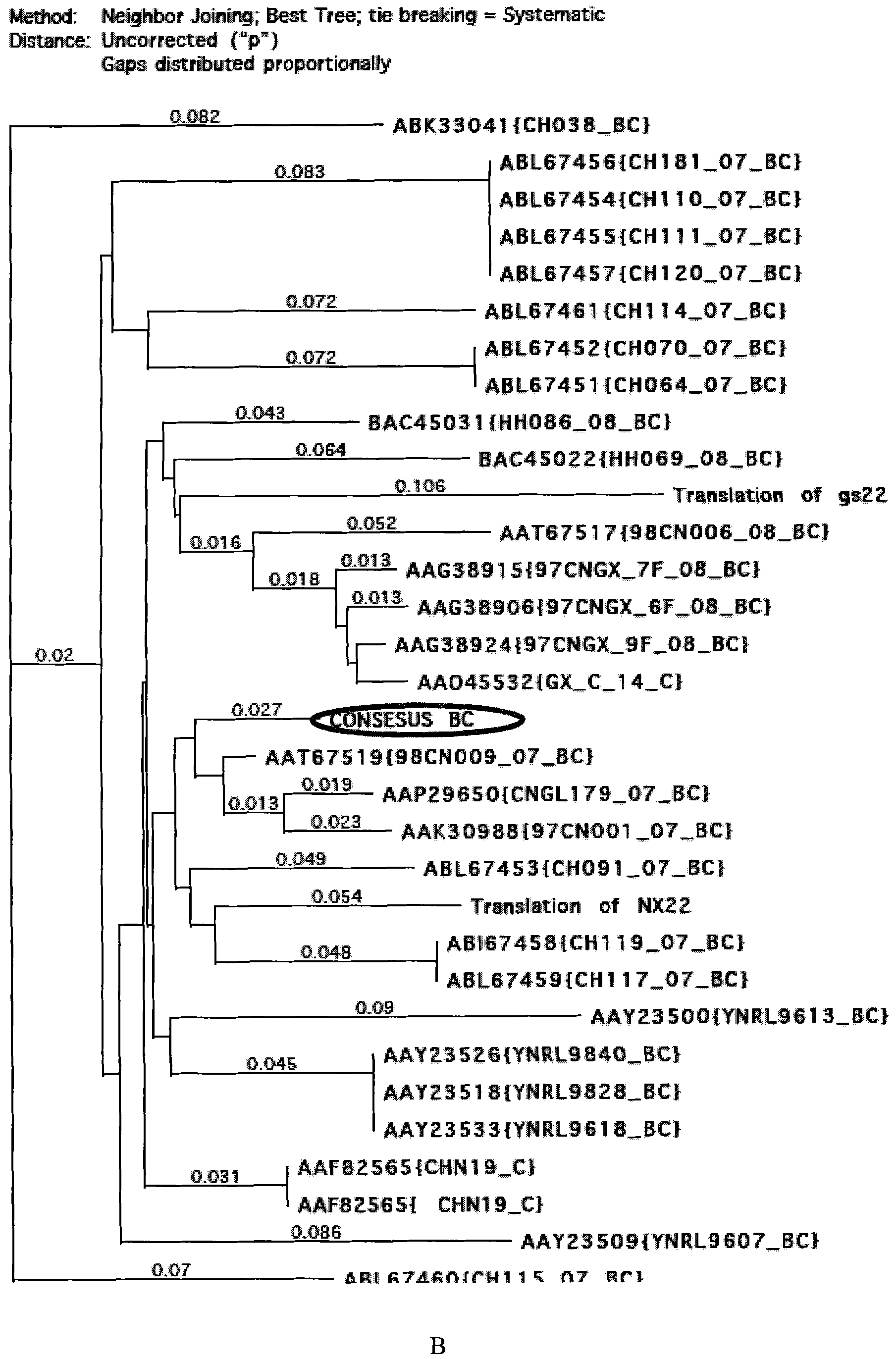
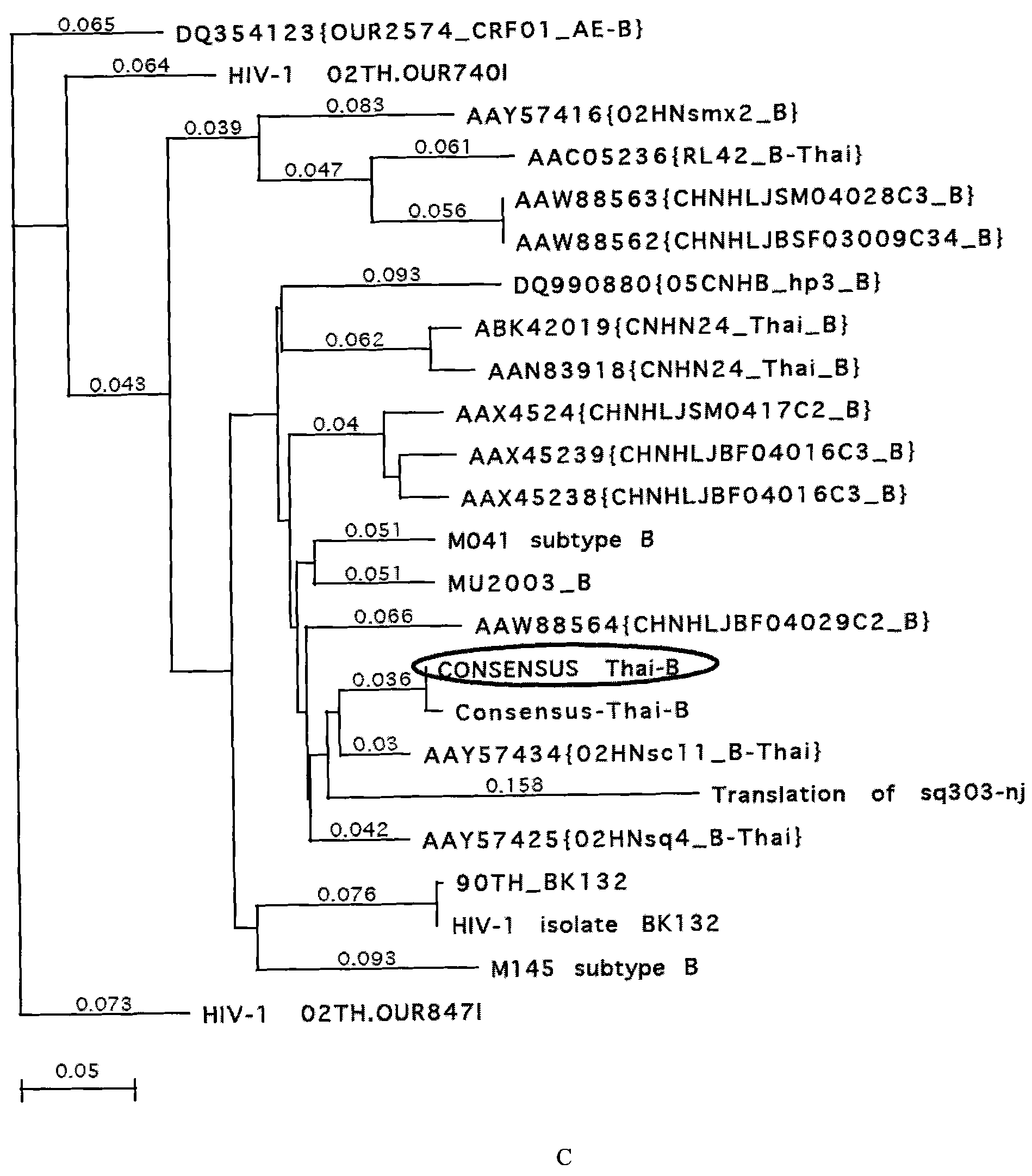
[0038] Example 1. Analysis of gp120 amino acid sequence of HIV-1AE, BC and ThB subtypes in China
[0039] The gp120 amino acid sequences of known Chinese HIV-1AE, BC and ThB subtypes in the gene bank were analyzed. According to the results of the sequence arrangement and the analysis results of the genetic evolution tree ( figure 1 ), designed the gp120 amino acid consensus sequences of Chinese HIV-1AE, BC and ThB subtypes.
Embodiment 2
[0040] Example 2. Design and synthesis of codons encoding consensus amino acid sequences
[0041] First, use the gene software MacVector 7.2 to analyze the gene sequence encoding the consensus amino acid sequence of the envelope protein gp120 of HIV-1 epidemic strains in China (A / E recombinant subtype, B / C recombinant subtype and ThB subtype), and find out its codons Bias and codon sites in the original sequence where codon usage bias differs from mammalian codon usage bias and from E. coli codon usage bias. For amino acids with the same codon usage preference in mammals and Escherichia coli, the codons encoding the amino acid in the original sequence were replaced by codons favored by both mammals and Escherichia coli, and codon-optimized coding AE, BC and ThB were designed respectively The gene sequences of subtype gp120 proteins are respectively SEQ ID NO.4, SEQ ID NO.5 and SEQ ID NO.6. After codon optimization, the DNA sequence of gp120 gene changed, but the amino acid se...
Embodiment 3
[0042] Example 3. gp120DNA vaccine construction
[0043] Plasmids pJW4303 and pGA4 / AE.gp120 (or pGA4 / BC.gp120, or pGA4 / ThB.gp120) were double digested with Nhe I and BamH I, respectively. The enzyme digestion reaction system is: 10×Buffer TangoTM 4 μL, 10×BSA 4 μL, plasmid 20 μL, NheI 1 μL, BamHI 1 μL, and water to 40 μL. After the digested product was subjected to 7g / L agarose gel electrophoresis, the target fragment of about 1400bp and the linearized pJW4303 were recovered and purified with a gel recovery kit (Agarose Gel DNA Purification Kit Ver.2.0, Dalian Bao Biological Co., Ltd.).
[0044] Place the agarose gel after electrophoresis under the ultraviolet detector, cut off the gel containing the target DNA, weigh the mass of the gel block with an analytical balance, and calculate the volume of the gel block according to 1 mg = 1 μL. Add 3 times the volume of DR-I Buffer, place in a 75°C water bath to heat and melt the gel block, mix well every 2 minutes until the gel blo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com