Real-time quantitative PCR detection method for red-sea bream iridovirus
A real-time quantitative technology of red sea bream iridescent virus, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, fluorescence/phosphorescence, etc. The effect of sample cross-contamination, reducing environmental pollution and wide detection range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
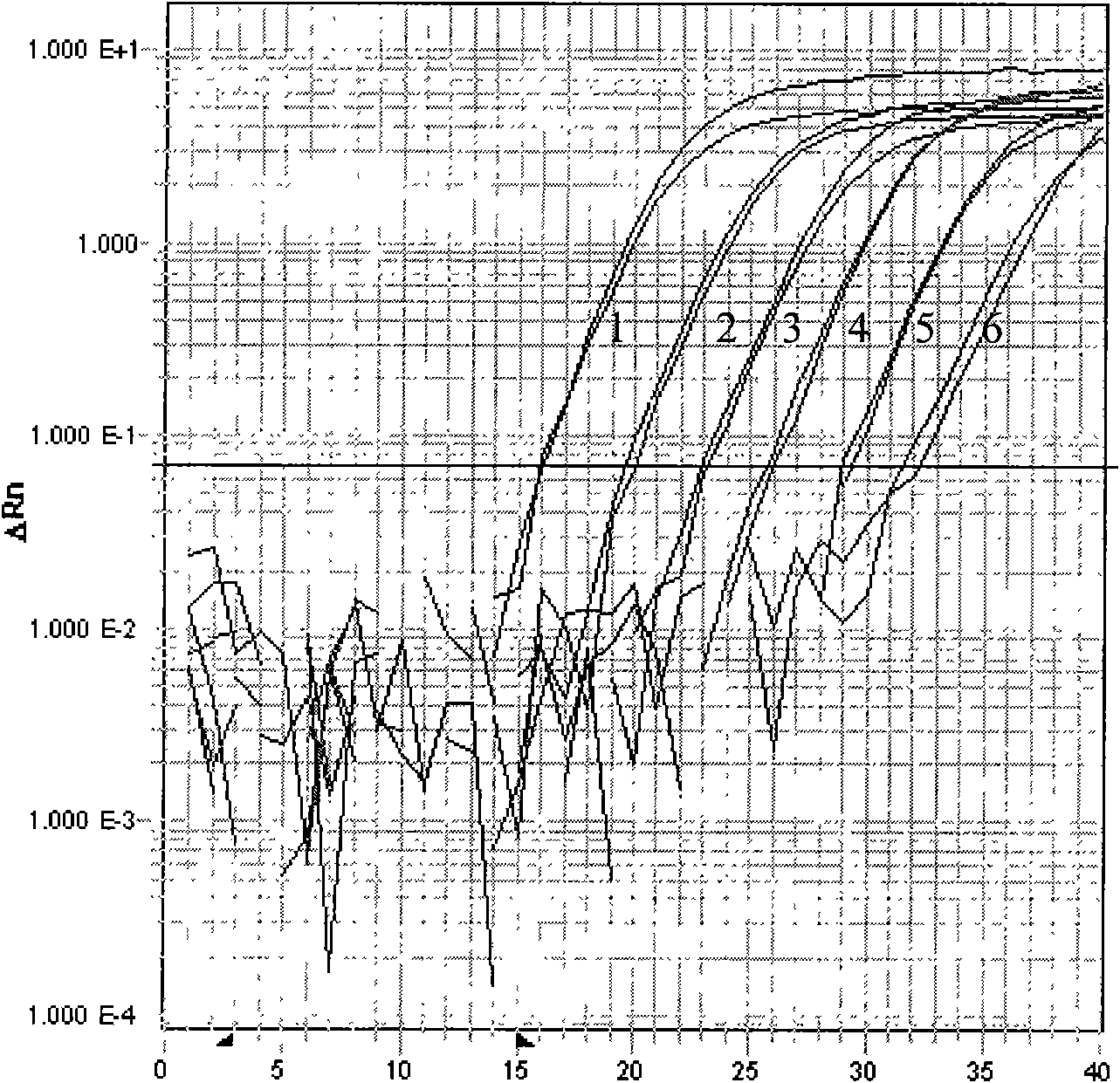
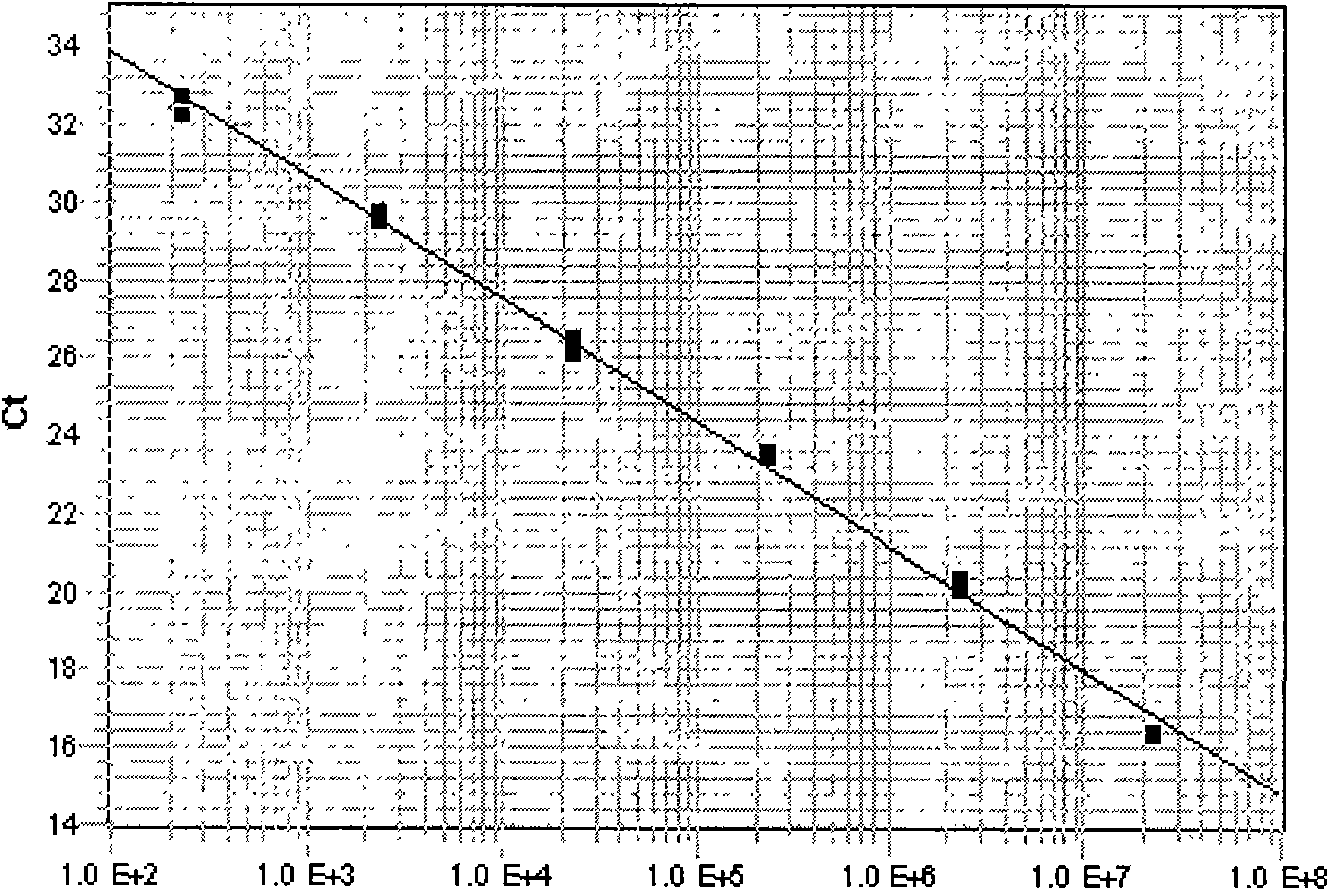
[0017] Taking perch samples as an example, real-time quantitative PCR method with fluorescent probes was used to quantitatively detect red sea bream iridescent virus; sea bass samples were collected from a farm in Shandong Province, and had typical clinical symptoms of red sea bream iridescent virus infection, which appeared on the skin, fins and eyeballs of the fish. Small vesicle-like swellings, with medical history of ciliates, tympanum, etc.; the above-mentioned designed specific primer sequences and fluorescent probe sequences:
[0018] In http: / / www.ncbi.nlm.nih.gov / Genbank / , search for the sequence of red snapper iridescent virus, and use the existing DNALASERGENE 7.1 software to analyze and compare the conservation of each gene sequence. The results show that the main red snapper iridescent virus is The capsid protein (MCP) gene has better conservation and research value, and is a major trend in virus research at present. Therefore, using the MCP gene as the target gen...
Embodiment 2
[0035] Taking the perch sample as an example, utilize SYBR GREEN I fluorescent dye to detect whether the perch sample contains red sea bream iridescent virus; Under the circumstances, there is no need to design a fluorescent probe; the real-time quantitative PCR reaction system and reaction procedure are the same as in Example 1, except that 1 μL of 10 μmol / L fluorescent probe in the real-time quantitative PCR reaction system is replaced with 1 μL of 25×SYBR GREEN I (ROCHE) fluorescent dye. Moreover, after the real-time quantitative PCR reaction program is completed, use the ABI 7900HT quantitative PCR instrument for melting curve analysis. The program is 95°C for 15 minutes, 60°C for 15 minutes, and 95°C for 15 minutes. The heating and cooling rate is 1°C per 25 seconds. Finally, the Tm value of the sample is given by the SDS 2.1 software; the instrument collects the fluorescence signal, and the analysis data is the same as in Example 1; the melting curve of the detection sam...
Embodiment 3
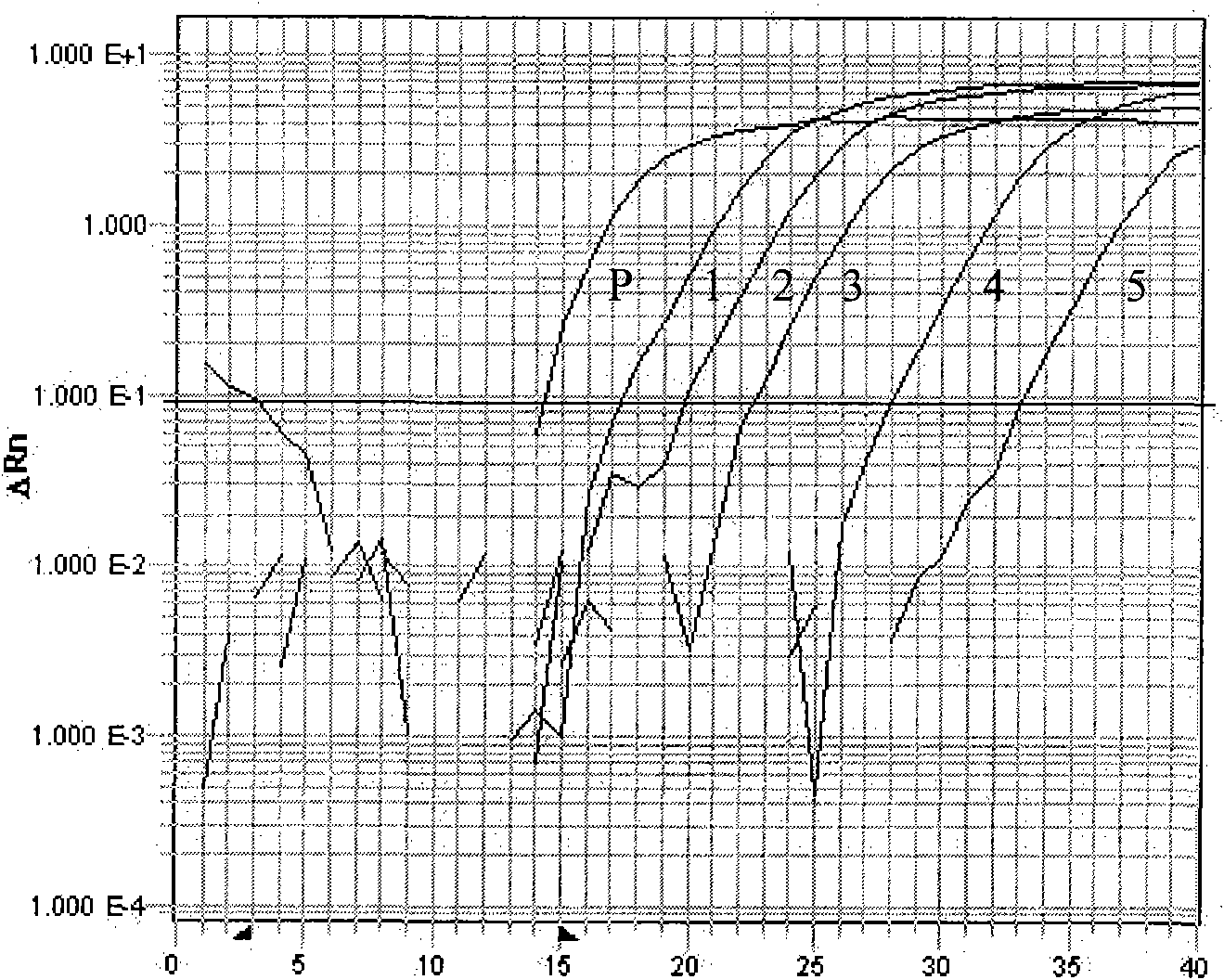
[0037] To analyze the specificity of SYBR GREEN I real-time quantitative PCR detection method for red sea bream iridescent virus; as a control, other viruses that are closely related to red sea bream iridescent virus were used for detection by SYBR GREEN I real-time quantitative PCR method, and these viruses belonged to Iridoviridae, they are epidemic hematopoietic necrosis virus (EHNV), lymphocyst virus (LCDV), frog virus 3 (FV 3), soft shell turtle iridescent virus (STIV); detection step is the same as embodiment 2, just will treat The DNA of the test sample was replaced with the DNA of epidemic hematopoietic necrosis virus (EHNV), lymphocyst virus (LCDV), frog iridescent virus (BIV), soft shell turtle iridescent virus (STIV); melting curve of SYBR GREEN I real-time quantitative PCR See Figure 5 Among them, the Tm value of the amplified product of red sea bream iridescent virus was 82.2°C, the Tm value of the other four viruses exceeded the range of 82.0°C-830, and the fluo...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com