Modified cyanobacteria
A technology of cyanobacteria and bacteria, applied in the field of bacteriology, can solve the problem of low light energy productivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0173] Modified Synechocystis PCC 6803 Cyanobacteria for Increased Lipid Content
[0174]Synechocystis PCC 6803, a cyanobacteria, was modified to overexpress the VIPP1 gene sll0617. To achieve high expression levels of the vippl gene (sll0617), it was cloned under the constitutive native promoter of the Synechocystis psbA3 gene (sll1867). The vippl gene was cloned between the upstream (287 bp) and downstream (394 bp) regions of the native psbA3 gene in the spectinomycin analog of the pA31hcgA3 plasmid (He et al., 1999); Figure 14 shown in. The sequence of the vippl (sll0617) gene was obtained from CyanoBase, and two primers were constructed to amplify the 804 bp vippl gene of the Synechocystis genome, 15 bp upstream of the vippl gene start codon, and 18 bp downstream of the vippl gene stop codon. 837bp fragment. The sequences of the primers were 5'-GAG GATAAG TAA GtC ATG aGA TTA TTT GAC and 5'-CTG GCT GAG TTA AtgCAt TTA CAG ATT ATT TAA CC. Lowercase letters indicate nucle...
Embodiment 2
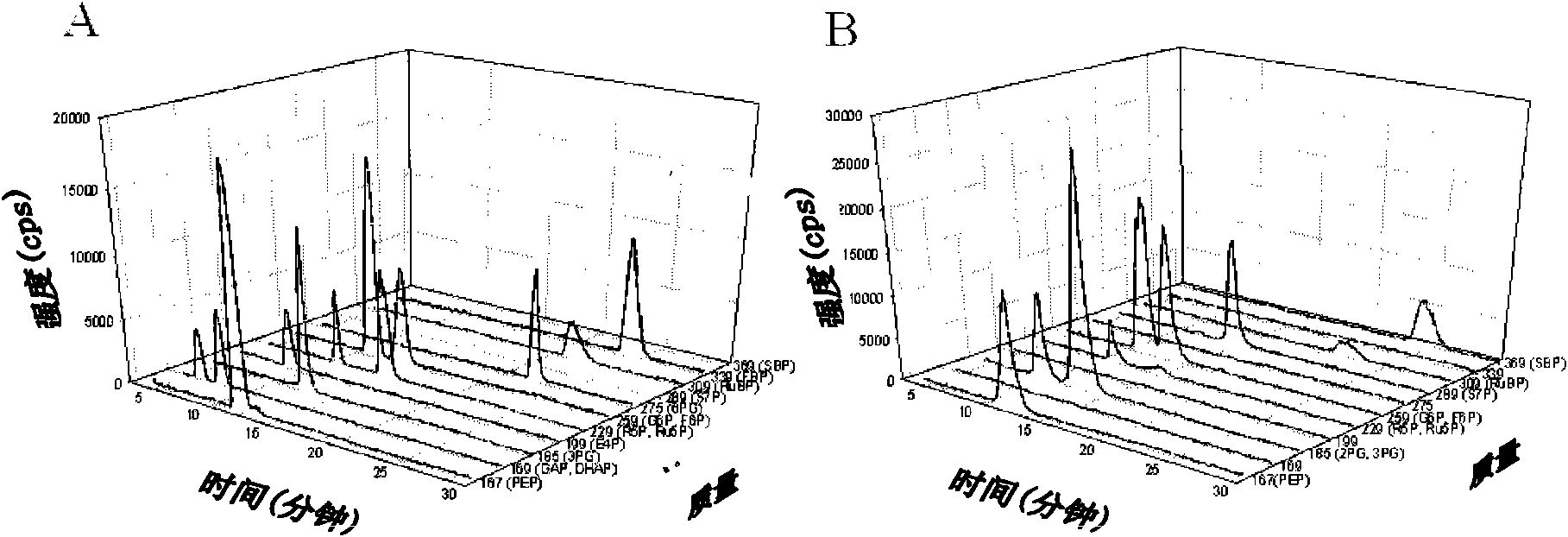
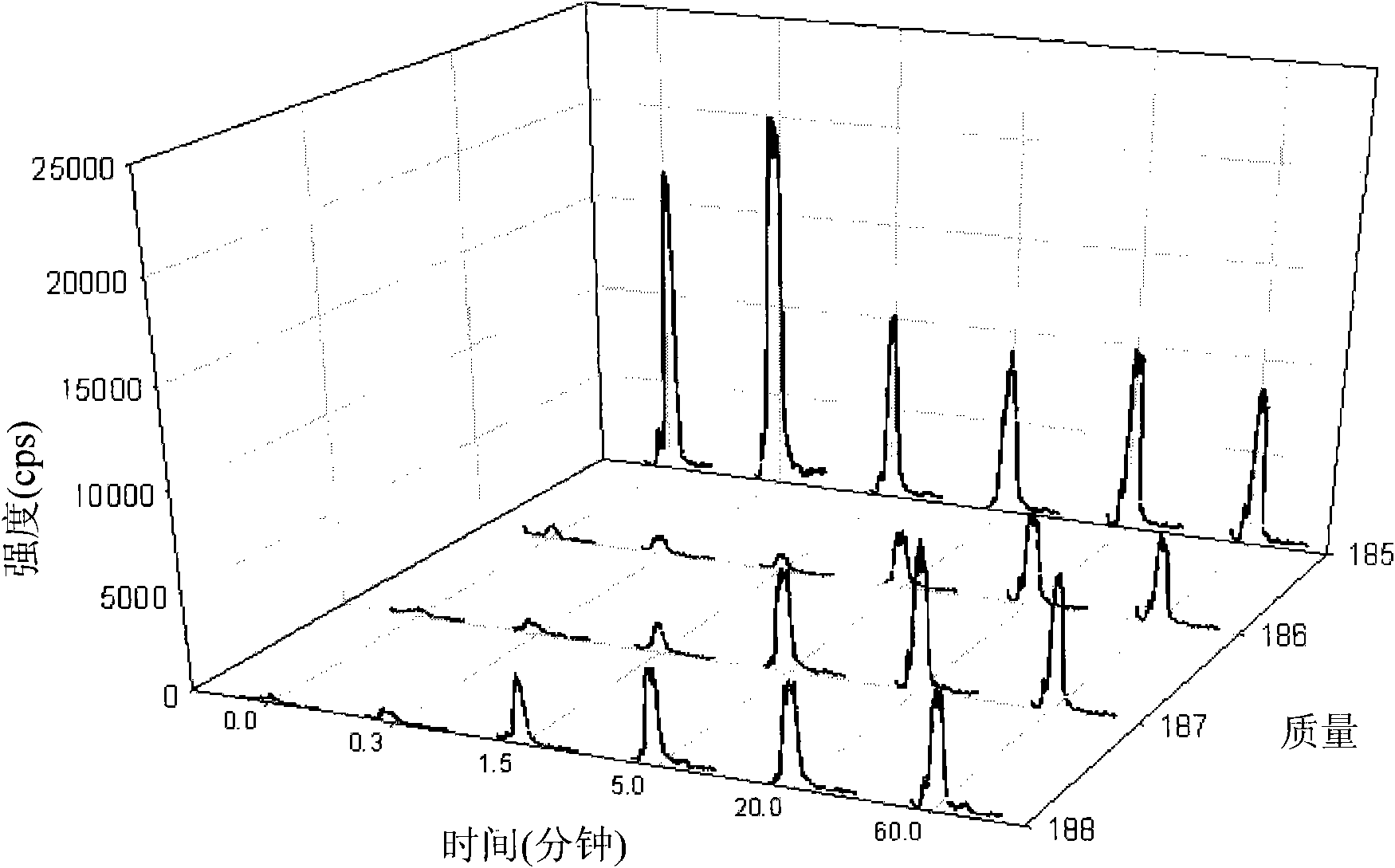
[0186] Dynamic analysis of metabolic flux through the central carbohydrate metabolic pathway of Synechocystis PCC 6803,
[0187] and enhancement of fatty acid biosynthesis by accABCD overexpression
[0188] 1. Materials and methods
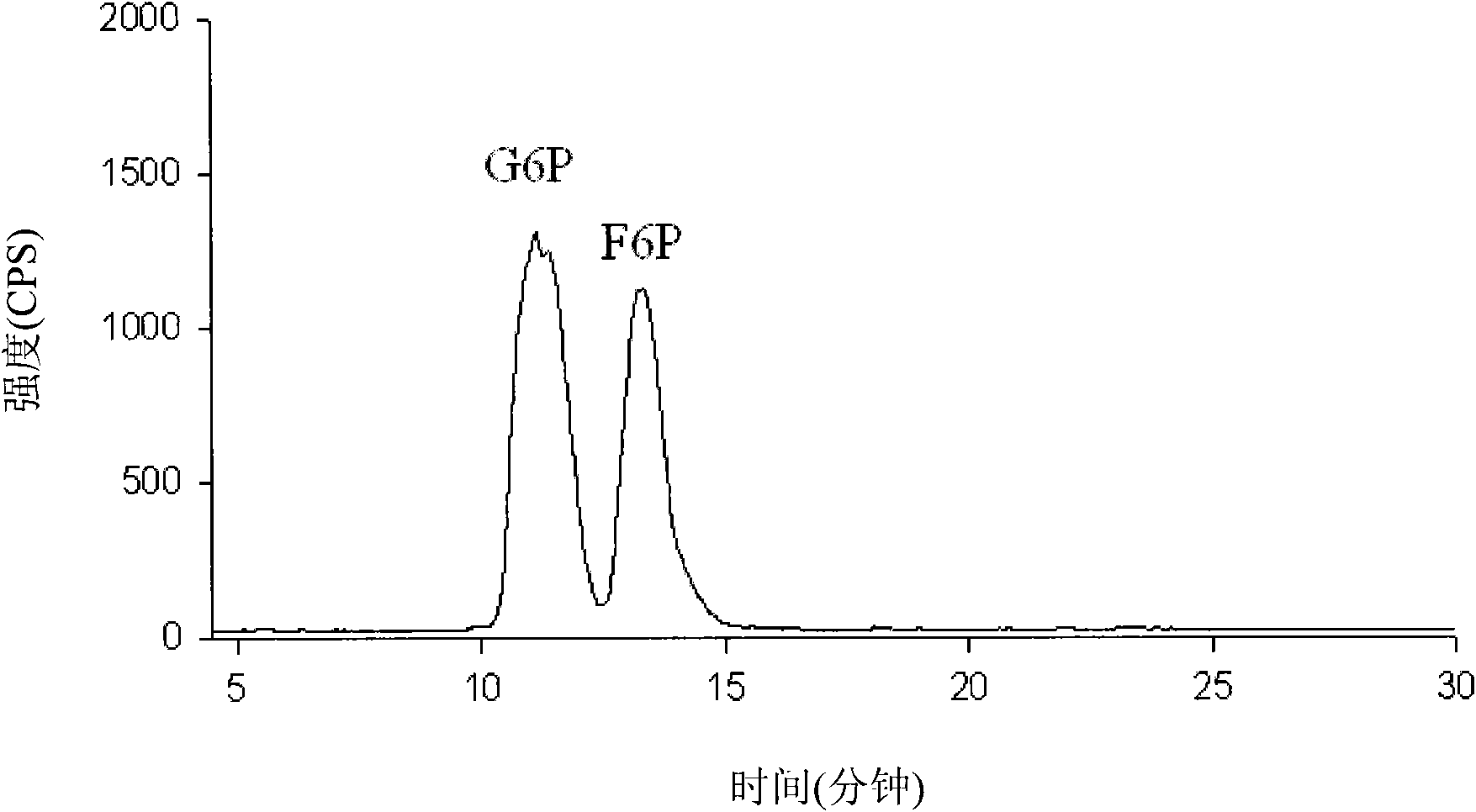
[0189] Chemical material. Chemicals used as standards (glucose-6-phosphate (G6P), fructose-6-phosphate (F6P), fructose-1,6-bisphosphate (FBP), glyceraldehyde-3-phosphate (GAP), diphosphate Hydroxyacetone (DHAP), 3-phosphoglycerate (3PG), phosphoenolpyruvate (PEP), 6-phosphogluconate (6PG), ribose-5-phosphate (R5P), ribulose-5-phosphate ( Ru5P), ribulose-1,5-bisphosphate (RuBP) and erythrose-4-phosphate (E4P)) were purchased from (St. Louis, MO). evenly 13 C-labeled D-glucose (U- 13 C 6 -D-glucose) was obtained from Cambridge Isotope Laboratories, Inc. (Andover, MA). Milli-Q grade water (Millipore, Van Nuys, CA) was used for all solutions.
[0190] growth conditions and 13 C-glucose label. Synechocystis sp. PCC6803 wild-type and mutants l...
Embodiment 3
[0257] Modified cyanobacteria for increased PHB (bioplastic) content
[0258] 1. Materials and Methods
[0259] Bacterial Strains and Culture Conditions: To understand the physiological role of PHB synthesis in Synechocystis sp. PCC 6803, a series of mutants with altered metabolic pathways resulting in significantly different PHB content were compared under different culture conditions. Deficiency of three terminal oxidases (cytochrome aa 3 mutants of cytochrome c oxidase (CtaI), putative cytochrome bo-type quinol oxidase (CtaII), and cytochrome bd-type quinol oxidase (Cyd)) (Howitt et al., 1998). PS II deletion / oxidase deletion mutants lacking photosynthetic oxygen release and respiratory oxygen consumption were then established by additional deletion of psbB encoding the CP47 protein of photosynthetic system II (Howitt et al., 2001). CyanoRubrum, a mutant strain donated by Dr. Michael Gurevitz (Tel Aviv University, Israel), in which the original cyanobacteria RuBisCO gene ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com