Recombinant adenovirus of Survivin promoter regulated human NIS gene, construction method and use
A technology for recombining adenoviruses and promoters, applied in the field of biomedicine, can solve the problems of weak starting ability and narrow range of promoters, etc., and achieve the effects of high recombination efficiency, simplified screening work, and simple operation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Survivin promoter PCR amplification:
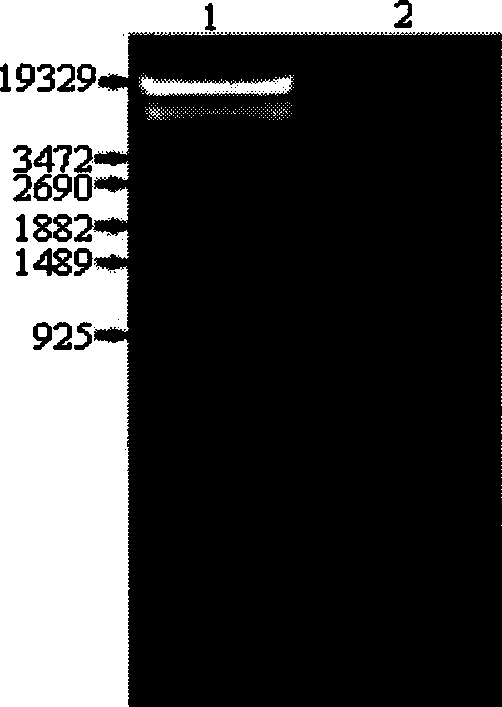
[0056] Primers were designed according to the sequence of Survivin DNA (NC 000017) in GeneBank, upstream primer: 5′- AGATCT GCACGCGTTCTTTGAAAGC-3'(-222~-204), downstream primer: 5'- AAGCTT CCACCTCTGCCAACGGGTC-3' (+21-+39), respectively containing BglII and HindIII restriction enzyme sites (synthesized by Shanghai Handsome Biotechnology Co., Ltd.). Using the genomic DNA of PC-3 cells as a template, the Survivin promoter fragment was amplified by PCR, pre-denatured at 95°C for 5 minutes, denatured at 95°C for 30s, annealed at 56°C for 30s, extended at 72°C for 30s, and after 25 cycles, extended at 72°C for 10 minutes. Gel recovery 262bp product fragment, such as figure 1 Shown, the second passage of agarose gel electrophoresis shows the Survivin promoter band that PCR amplification obtains, and size is 262bp; T / A clones to PMD-18T vector (Japan Takara Company), named after PMD-Sur, sequencing (Completed by Shanghai Handsome Biot...
Embodiment 2
[0058] Construction of the luciferase reporter plasmid vector pGL3-Sur-Luc driven by the Survivin promoter, and determination of transcriptional activity:
[0059] PMD-Sur and pGL3-Basic plasmids (promega, USA) were digested with BglI and HindIII, 262bp and 4.8kb fragments were recovered by gel, ligated and transformed into JM109 competent cells, positive clones were selected and named pGL3-Sur-Luc. like figure 1 As shown, the third lane of agarose gel electrophoresis shows the 4.8kb and 262bp bands obtained after pGL3-Sur-Luc was cut with BglII and HindIII.
[0060] Lipofectamine TM In 2000, pGL3-Sur-Luc, pGL3-SV40-Luc, pGL3-Basic and the internal reference plasmid pRL-CMV (the latter three are products of Promega, USA) were co-transfected into the human hormone-dependent prostate cancer cell line LNCaP, human Hormone-independent prostate cancer cell lines DU145 and PC-3, human liver cancer cell HepG2, malignant melanoma cell A375 and normal human dental pulp fibroblast DPF...
Embodiment 3
[0062] Amplification of human NIS gene
[0063] Extract the total RNA of human thyroid tissue according to the operation guide of the kit, synthesize the first strand of cDNA by reverse transcription, and then use this as a template to PCR amplify the codable region of human NIS gene. The primer sequence is NISP1:CCCAAGCTTATGGAGGCCGTGGAGACCGGGGAAC(348-372) NISP2: GCGTCTAGATCAGAGGTTTGTCTCCTGCTG (2259-2280), in which restriction endonuclease HindIII and XbaI sites are added to the 5' ends of the P1 and P2 primers, respectively. The reaction conditions were pre-denaturation at 95°C for 5 min, denaturation at 94°C for 45 s, annealing at 70°C for 30 s, decreasing by 0.5°C per cycle, extension at 72°C for 2 min, and extension for 5 s per cycle, a total of 20 cycles, denaturation at 94°C for 45 s, annealing at 60°C for 45 s, Extend at 72°C for 2.5min, and extend for 5s per cycle. After 15 cycles, extend at 72°C for 10min. Amplified fragments, such as figure 2 As shown, the 1.9Kb b...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Titer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com