Process for ultra-sensitive quantification of target analytes in complex biological systems
a biological system and target analyte technology, applied in the field of analytical separation and measurement systems and processes, can solve the problems of high failure rate, long development lead time, high cost of de novo development of specific antibodies for new immunoassays, etc., and achieve the effect of improving srm sensitivity and quantitation accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Blood Plasma and Serum Depletion
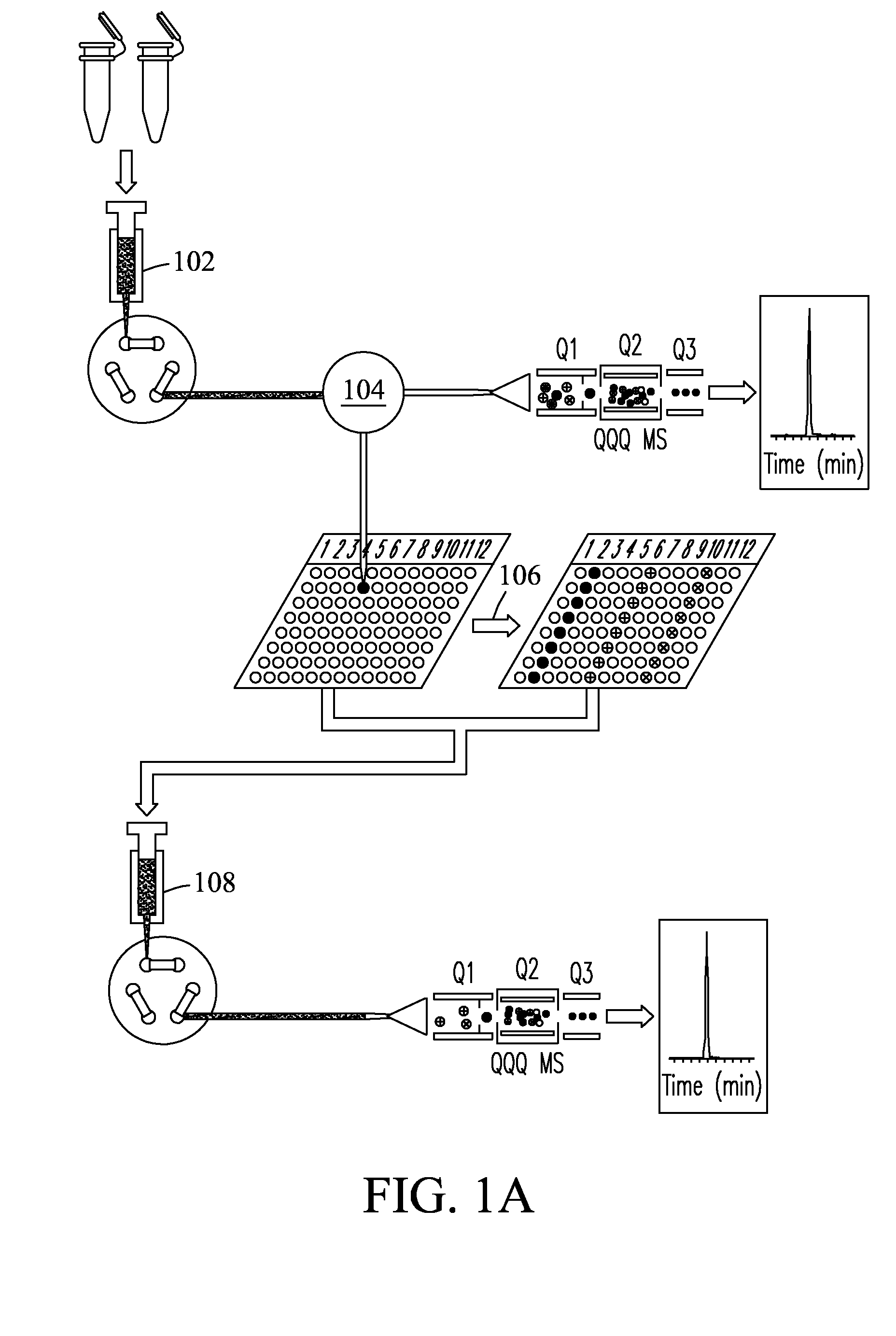
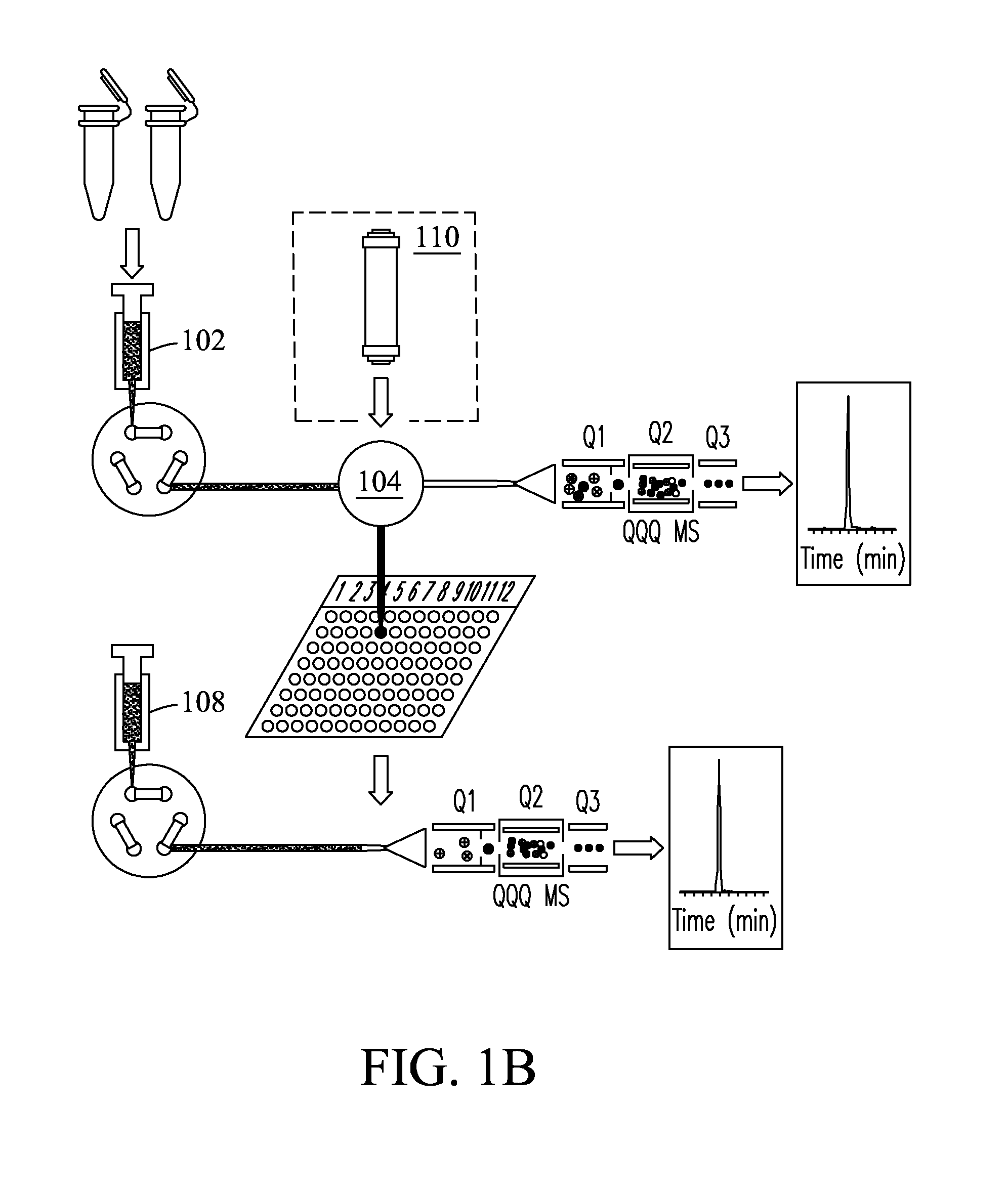
[0074]Fourteen high abundance plasma proteins (albumin; IgG; α1-antitrypsin; IgA; IgM; transferrin; haptoglobin; α1-acid glycoprotein; α2-macroglobulin; apolipoprotein A-I; apolipoprotein A-II; fibrinogen; complement C3; and apolipoprotein B) that constitute ˜95% of the total protein mass of human plasma were depleted from plasma and serum samples using a Seppro® IgY-14 LC10 immunoaffinity column (Sigma-Aldrich, St. Louis, Mo., USA) on an Agilent 1100 series HPLC system (Agilent, Palo Alto, Calif., USA). For each depletion experiment, ˜125 μL plasma / serum was diluted 10-fold and injected onto the IgY14 column. Following the IgY14 separations, the flow-through fraction was concentrated using Amicon® Ultra-15 (5 kDa nominal molecular weight limit, Millipore, Billerica, Mass., USA). Protein concentrations were determined by BCA protein assay (Pierce, Rockford, Ill.).
example 2
Protein Digestion and Spike-in Experiments
[0075]Protein samples from plasma / serum and cancer cells were digested with the same protocol. For the plasma spike-in experiments at the peptide level, IgY14 flow-through proteins from 10 injections were pooled, denatured, and then reduced using 8 M urea and 10 mM dithiothreitol (DTT) in 50 mM NH4HCO3 buffer (pH 8.0) for 1 h at 37° C. Samples were then alkylated using 40 mM iodoacetamide for 1 h at room temperature. The resulting protein mixture was diluted 6-fold with 50 mM NH4HCO3, after which sequencing grade modified porcine trypsin (Promega, Madison, Wis.) was added at a trypsin:protein ratio of 1:50 (w / w), and the sample digested at 37° C. for 3 hrs. The protein digest was subsequently cleaned up using an SPE_C-18 column (Supelco, Bellefonte, Pa.). The final peptide samples were stored at −80° C. until further usage. Proteins from plasma / serum and cancer cells samples were digested with the same protocol. Stocks of 1 μg / μL of each of ...
example 3
SRM Assay Configuration
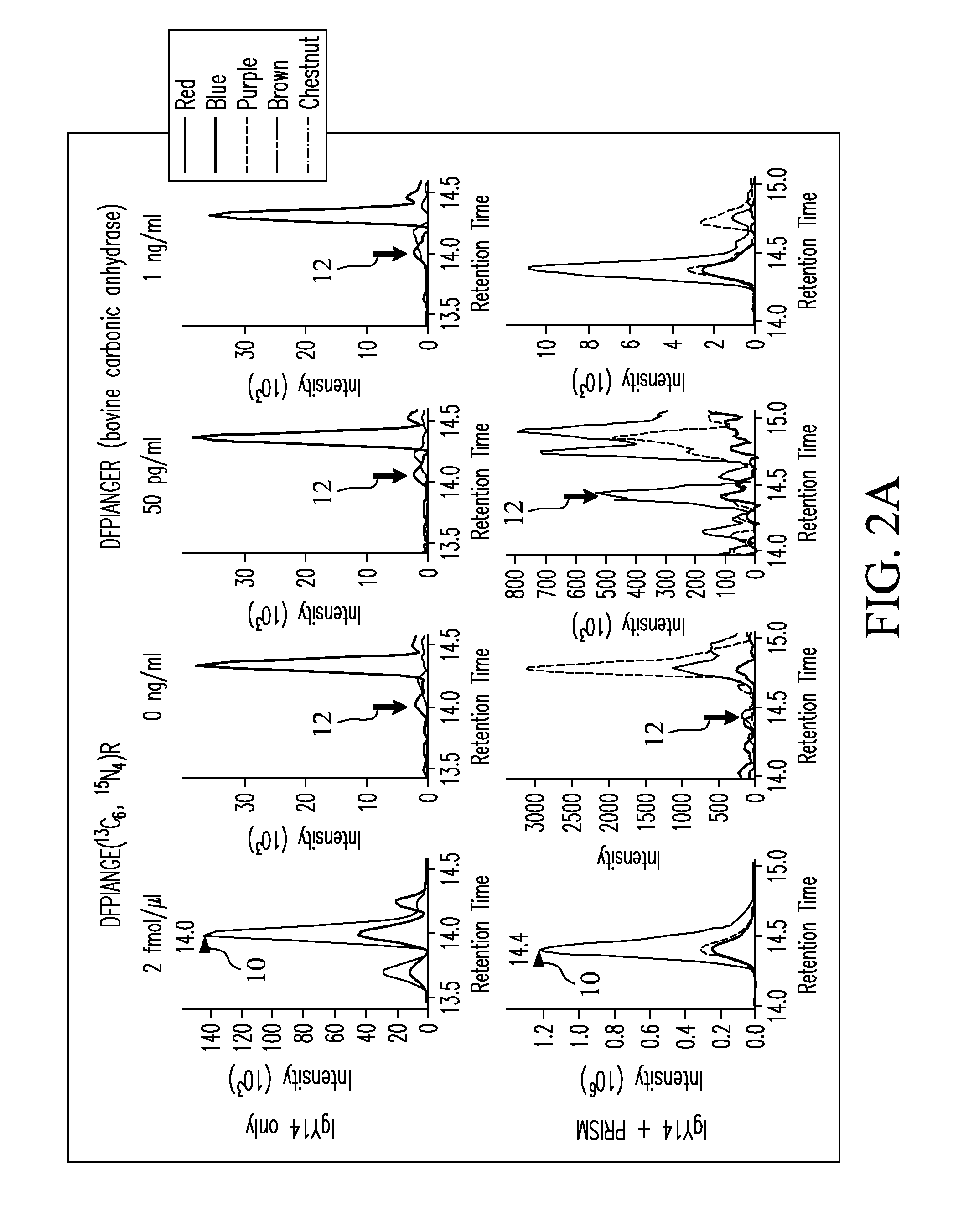
[0076]SRM assays were configured based on experimental tandem MS / MS data for peptide mixtures from four target proteins: 1) bovine carbonic anhydrase, 2) bovine β-lactoglobulin, 3) E. coli β-galactosidase, and 4) human prostate-specific antigen (PSA) (Sigma-Aldrich, St. Louis, Mo., USA). MS / MS spectra were used to select transitions by choosing the eight most intense y-type ions for each peptide of precursor charge state +2 or +3. For each target protein, two unique peptides were selected that had the best ionization and most intense fragmentation patterns when transmitted to the mass spectrometer. Precursor-to-fragment ion transitions (TABLE 1) were tested by SRM measurements on a TSQ Quantum Ultra triple quadrupole mass spectrometer (Thermo Fisher Scientific, Inc., Waltham, Mass., USA). Optimization of collision energy (CE) was performed by direct infusion of a 500 nM digested solution of each target protein dissolved in 0.1% formic acid in water at an infus...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com