Methods for treatment of bacterial infections
a technology of bacterial infections and dtranspeptidases, which is applied in the direction of antibacterial agents, biocide, heterocyclic compound active ingredients, etc., can solve the problems of reducing our ability to respond effectively to this threat, reducing the morphology of the colony, and reducing the virulence of the infection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
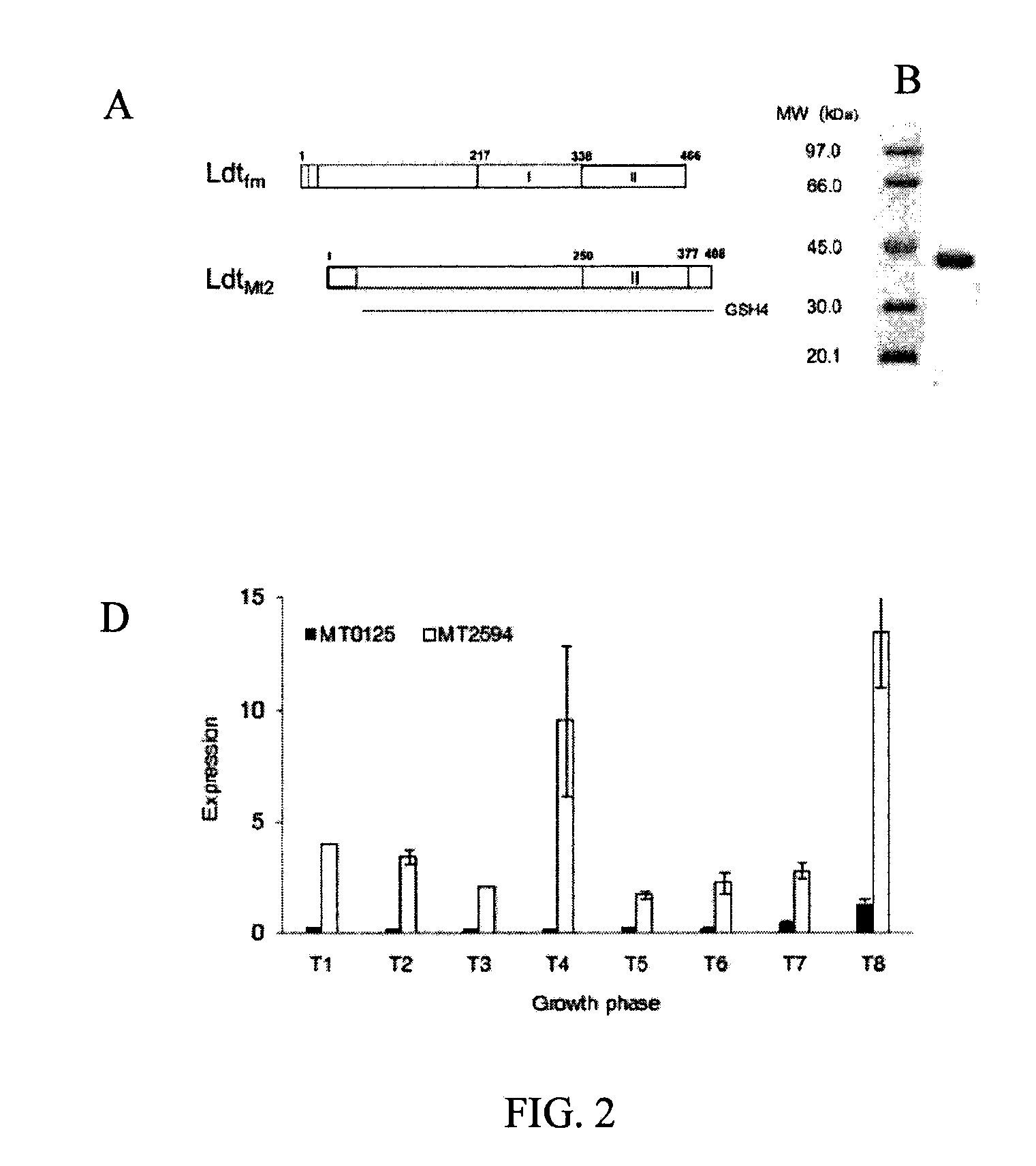
Production and Purification of Recombinant LdtMt2
[0035]A portion of the ltdMt2 gene was amplified with primers 5′-TTTTCATGAT-CGCCGATCTGCTGGTGC-3′ and 5′-TTGGATCCCGCCTTGGCGTTACCGGC-3′, digested with BspHI plus BamHI (underlined) and cloned into pET2818. The resulting plasmid, encodes a fusion protein consisting of a methionine specified by the ATG initiation codon of pET2818, residues 55 to 408 of LdtMt2, and a C-terminal polyhistidine tag with the sequence GSH6. E. coli BL21 (DE3) harboring pREP4GroESL and pET2818ΩldtMt2 was grown at 37° C. to an OD600 of 0.5 in 3 L of brain heart infusion broth containing 150 μg / ml ampicillin. Expression was induced by adding Isopropyl-D-thiogalactopyranoside to a final concentration of 0.5 mM and incubating the cultures for 17 hours at 16° C. LdtMt2 was purified from a clarified lysate by affinity chromatography on Ni2+-nitrilotriacetate-agarose resin (Qiagen GmbH, Germany) followed by anion exchange chromatography (MonoQ HR5 / 5, Amersham Pharmaci...
example 2
L, D-Transpeptidase Assays
[0036]We purified disaccharide-tetrapeptide containing amidated meso-diaminopimelic acid (GlcNAc-MurNAc-L-Ala1-D-iGln2-mesoDapNH23-D-Ala4) from C. jeikeium strain CIP103337 and determined the concentration after acid hydrolysis. In vitro formation of muropeptide dimers was tested in 10 μL of 50 mM Tris-HCl (pH 7.5) containing 300 mM NaCl, 5 μM LdtMt2, and 280 μM disaccharide-tetrapeptide. The reaction mixture was incubated for two hours at 37° C. the resulting muropeptides were analyzed by nanoelectrospray tandem mass spectrometry using N2 as the collision gas.
example 3
Bacterial Strains and Culture Conditions
[0037]M. tuberculosis CDC1551, a clinical isolate, was used as the host strain and a transposon insertion mutant in MT2594 (ldtMt2::Tn) was generated, isolated and characterized as described in Lamichhane et al., PNAS, 2003, 100(12): 7213-7218. This mutant carries a Himar1 transposon insertion at +872 base from the putative translation start site of the gene. The complemented strain was generated by transforming the mutant with pGS202—2594. This is a single copy integrating plasmid based on pMH94 backbone, which was modified into a GATEWAY compatible destination vector (Invitrogen) by inserting attL1 and attL2 sites. A wild-type copy of MT2594 along with its promoter was cloned into this destination vector pGS202 to generate pGS202—2594. Genotypes of the strains were verified by Southern blotting. For in vitro growth, Middlebrook 7H9 liquid medium supplemented with 0.2% glycerol, 0.05% Tween-80, 10% vol / vol oleic acid-albumin-dextrose-catalase...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Cell angle | aaaaa | aaaaa |
| Strain point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com