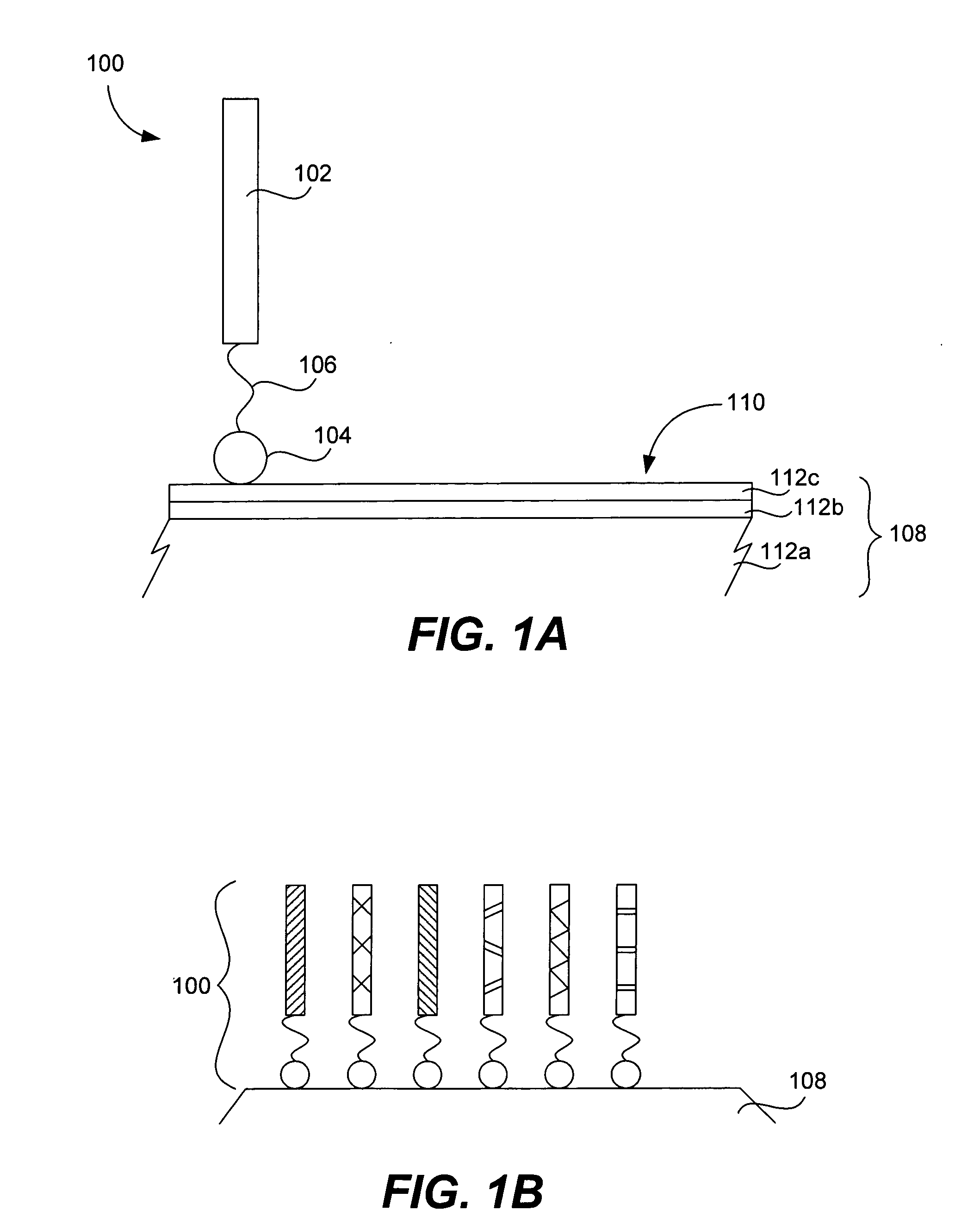
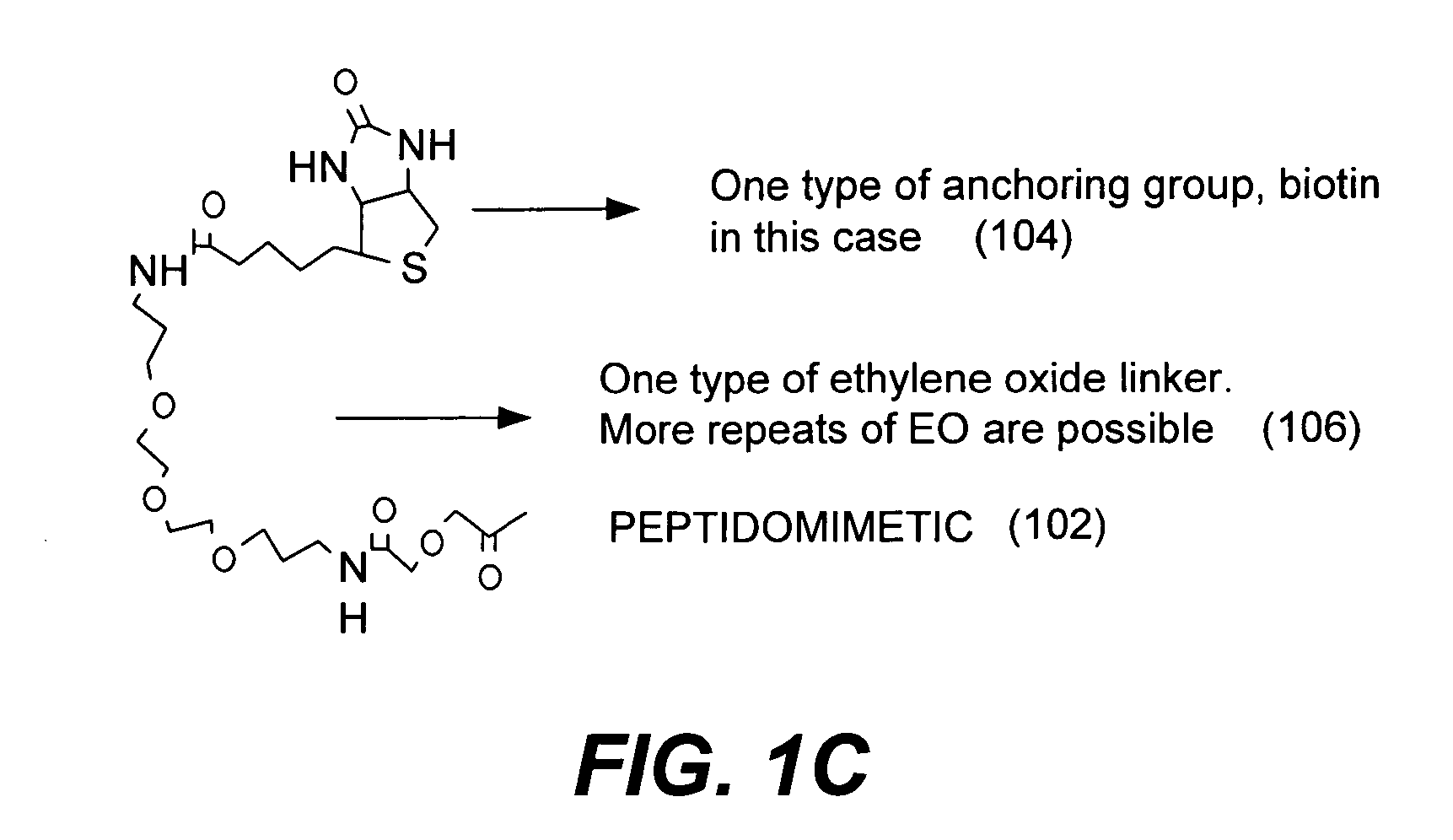
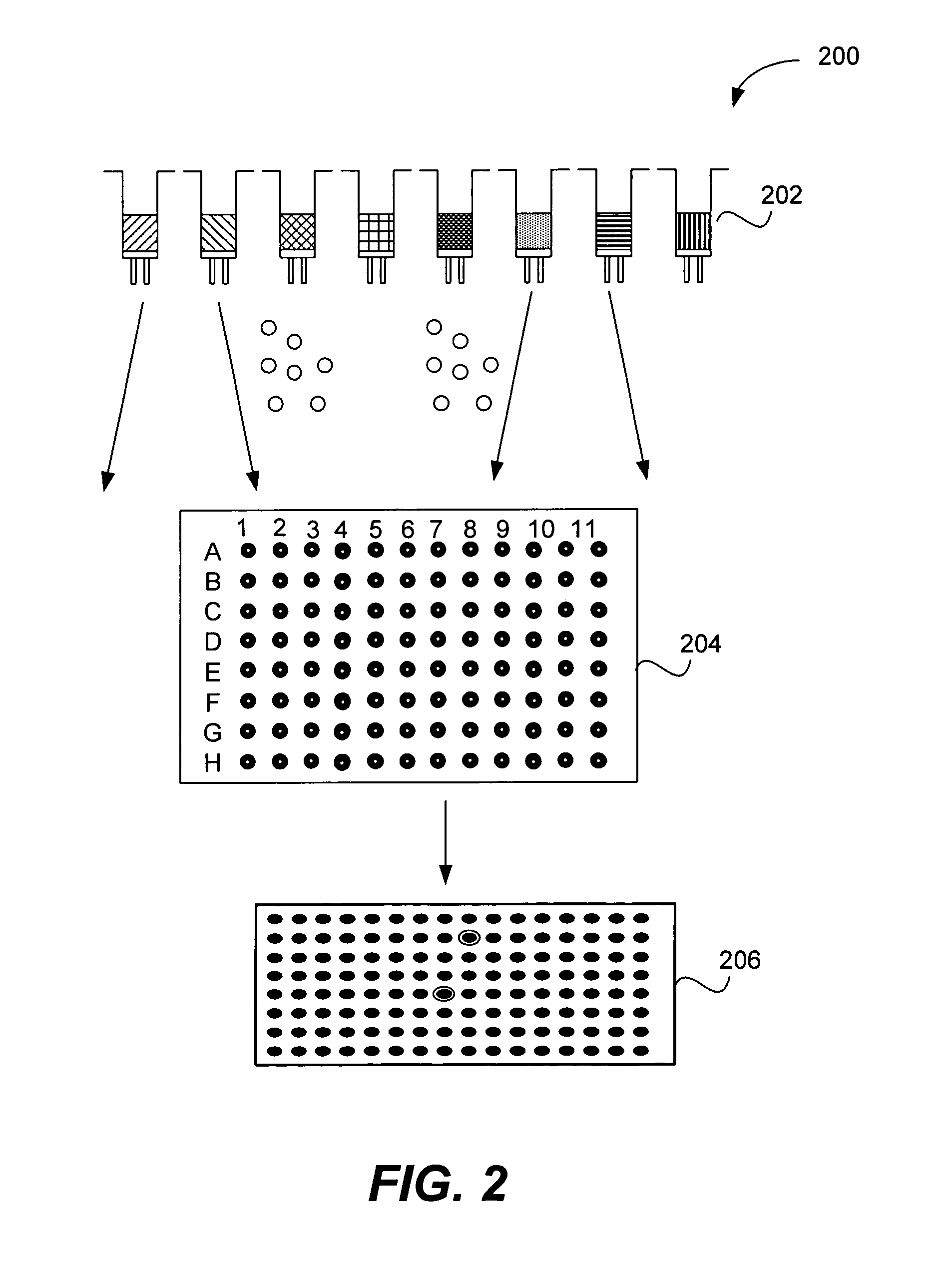
Microarrays on mirrored substrates for performing proteomic analyses
a proteomic analysis and microarray technology, applied in the field of cell product analysis and materials, can solve the problems of difficult to get reproducible data from 2-d gels, difficult to use expensive custom-made masks, and difficult to use custom-made masks. to achieve the effect of preventing non-specific binding
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of Microtiter Plates Containing One Compound Per Well
[0137] Preparation of one (1) 96-well plate containing 96 single compounds (0.8 mM) in a 1:1 DMSO / PBS solution: A siliconized vial (8 ml, 2 dram) was loaded with a one compound / bead library synthesized on Rink polystyrene macro-bead solid support (66 mg, ˜1320 beads, 40 nanomole / bead, 0.75 mmol / g, 425-500 um, Polymer Laboratories) that contains a total of 121 possible compounds. The beads were swollen in dichloroethane (DCE, 4 ml) for 24 hours and sieved over stainless steel mesh (600 μm). Using the resulting dichloroethane bead slurry, 96 beads were individually picked and relocated into a polypropylene 96-well plate (200 μL, conical bottom) in order to obtain one bead per well over 96 wells. The resulting 96-well plate, (the ‘grandmother’ plate), was transferred to a speed-vac evaporator Savant AES200 equipped with a 96-well plate carrier rotor and the remaining DCE was removed from the plate. A cleavage cocktail (T...
example 2
Functionalization of Solid Supports for Spotting
[0140] Gold or aluminum reflective microscope slides were used as substrates for spotting a library of peptoids. In one example, the peptoid is functionalized with a thiol endgroup and the surface is functionalized with a maleimide. Upon spotting, the thiol on the peptoid reacts with the maleimide on the surface to form a covalent thioether attachment. Gold surfaces activated with maleimide were prepared as follows: 1) Gold-coated microscope slides (1200 Angstroms Au, 30-50 Angstroms Ti or Cr) were cleaned with Chromic acid cleaning solution for 15 minutes and rinsed with HPLC grade water. 2) Gold slides were dipped into 1-5 mM amino-modified thiol (1-Mercaptoundecyldiethoxyamine; a C-11 alkyl, two ethylene oxides, and an amine; alternatively, C-2 to C-20 alkyl groups and / or ethoxy or triethoxy groups could be used in such a compound) for 1-24 hours at room temperature or at 45 or 60° C. The slides were rinsed four times in absolute e...
example 3
Derivatization of Aluminum / SiO2 Surfaces with Avidin
[0143] Aluminum slides coated with aminosilane were dipped into a solution of NHS-LC-LC-biotin (“LC” refers to 6-aminohexanoyl and “NHS” refers to N-hydroxysuccinimidyl) (commercially available from Pierce), depicted in FIG. 7A, that was 0.39 mM in PBS buffer. The slides were coated for 1.5 hours with shaking at 80 rpm. After attachment of biotin, the slides were rinsed with water, then dipped in a solution of 1 ug / ml-1 mg / ml avidin, streptavidin or neutravidin in PBS buffer for 2 hours, stirring at 70 rpm. The slides were rinsed with water and ready for spotting biotinylated peptides or peptoids. FIG. 7B depicts a representation of an avidin-derviatized aluminum slide spotted with a biotinylated protein-binding agent in accordance with one embodiment of the present invention.
[0144] The various surface modification steps was followed using ellipsometry to note the thickness changes. A thickness change increase of 40-45 angstroms ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thick | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com