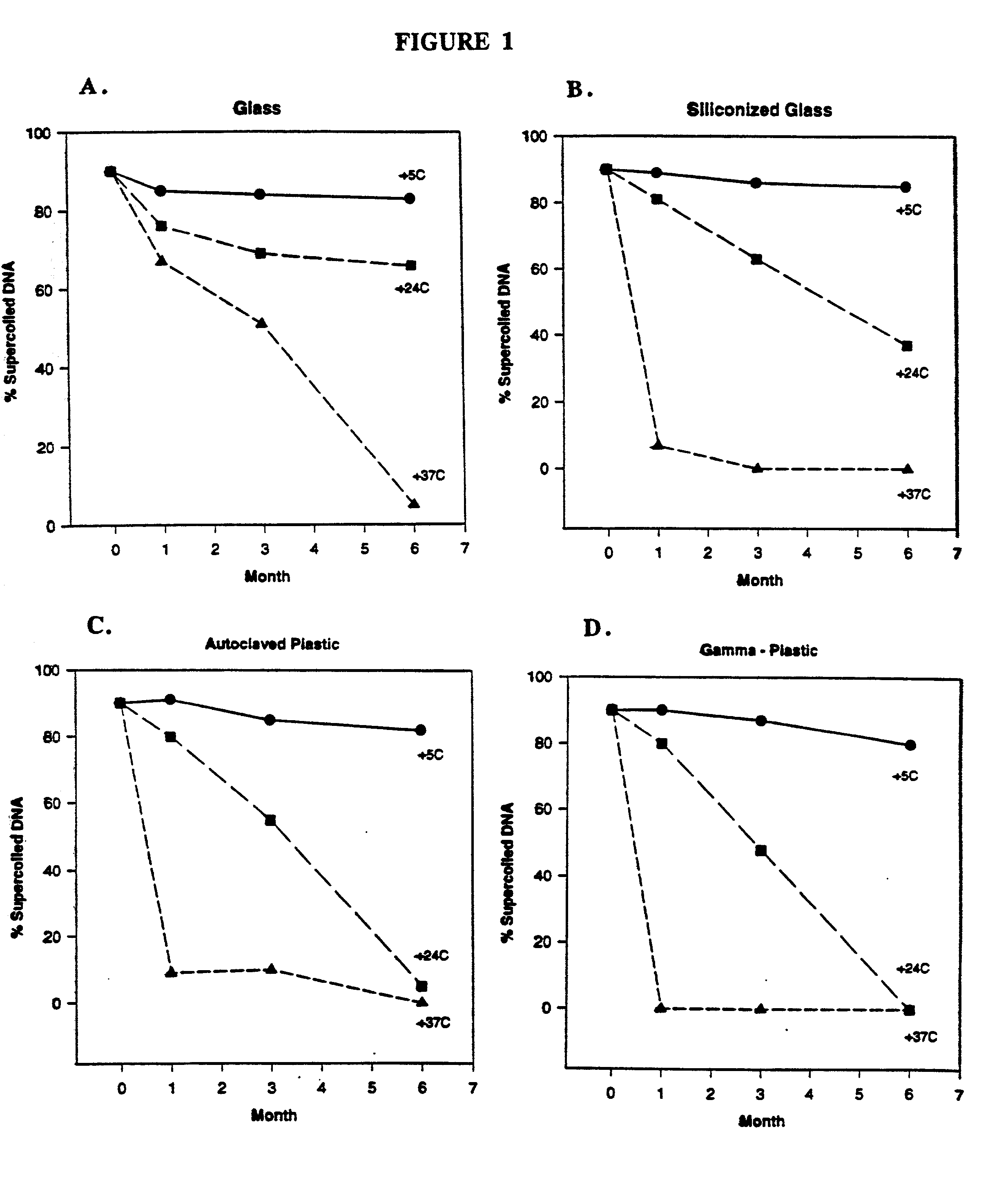
DNA vaccine formulations
a technology of nucleic acid and vaccine, applied in the field of new formulations of nucleic acid pharmaceutical products, can solve the problems of significant enhancement of stability, no increase in stability, dna degradation, etc., and achieve the effects of improving stability, improving stability, and increasing dna stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 2
[0089] Influenza Virus Gene Constructs In Expression Vector V1j
[0090] Many of the genes from the A / PR / 8 / 34 strain of influenza virus were cloned into the expression vector V1J, which gives rise to expression at levels as high or higher than in the V1 vector. The PR8 gene sequences are known and available in the GENBANK database. For each of the genes cloned below, the size of the fragment cloned was checked by sizing gel, and the GENBANK accession number against which partial sequence was compared are provided. For a method of obtaining these genes from virus strains, for example from virus obtained from the ATCC (A / PR / 8 / 34 is ATCC VR-95; many other strains are also on deposit with the ATCC).
[0091] A. Subcloning the PR8 Genes into V1J:
[0092] 1. NP
[0093] The NP gene was subcloned from pAPR501 (J. F. Young, U. Desselberber, P. Graves, P. Palese, A. Shatzman, and M. Rosenberg (1983), in The Origins of Pandemic Influenza Viruses, ed. W. G. Layer, (Elsevier, Amsterdam) pp.129-138). It wa...
example 3
[0110] Growth of Microbial Cells, Cell Lysis and Clarification
[0111] One liter of frozen E. coli cell slurry was used to make 8 liters of cell suspension in STET buffer (8% sucrose, 0.5% TRITON, 50 mM TRIS buffer, pH 8.5 and 50 mM EDTA). The absorbance of the cell suspension at 600 nm was about O.D. 30. The suspension was stirred continuously to ensure homogeneity. The viscosity of the cell suspension was measured to be about 1.94 cp at room temperature (24.degree. C.). The cell suspension was pumped through the heat exchanger at 81 mL / min which corresponded to a residence time of the cell solution in the heat exchanger of about 35 seconds. The bath temperature was maintained at 92.degree. C. The inlet and outlet temperatures of the cell solution were measured to be about 24.degree. C. and about 89.degree. C. (average), respectively. About 1 liter of sample was run through the heat exchanger. No visible clogging of the tube was observed although the lysate was somewhat thicker than ...
example 4
[0113] Control and Reproducibility of Cell Lysis with the Heat Exchanger
[0114] Adjusting the flowrate (i.e., residence time) at which the cell slurry is pumped through the heat exchanger permits tight control of the temperature of lysis, i.e., the outlet temperature. A cell slurry solution was prepared as described in Example 3 and pumped through the heat exchanger at flow rates ranging from 160 to 850 mL / min. The corresponding outlet temperatures ranged between 93.degree. C. and 65.degree. C., respectively. The initial temperature of the cell slurry was 24.degree. C. and the bath temperature was kept constant at 96.degree. C. In addition, a number of runs were performed where an outlet temperature of 80.degree. C. was targeted. Yields of 24 mg of circular DNA per L of clarified supernatant were consistently obtained demonstrating the reproducibility of the process.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com