Surface glucoprotein gp160 of recombination expression human acquired immunity defact virus 1
A surface glycoprotein and expression cassette technology, applied in the field of bioengineering, can solve problems such as high or low expression levels
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Obtaining HIV-1 gp160 gene encoded by 6 histidines at the C-terminal by PCR method
[0046] The DNA plasmid pN2gpt-gp160 (US Patent No. 5,445,953) carrying the HIV-1 gp160 gene was used as a template, and the following pair of primers were used to carry out conventional DNA polymerase chain reaction (PCR) under the action of polymerase.
[0047] 5' end primer:
[0048] 5'GCT CTA GAG AGC AGA AGA CAG TGG CAA TG 3' (SEQ ID NO: 3)
[0049] 3' end primer:
[0050] 5’CGA ATT CTA TTA GTG GTG ATG GTG ATG GTG TAG CAAAAT CCT TTC CAA GCC CTG 3' (SEQ ID NO: 4, the underlined part corresponds to 6His)
[0051] The resulting amplified product was digested with XbaI and inserted into the working plasmid pUC19 (purchased from New England Biolab) that had been digested with XbaI and blunt-ended at the HidIII site. Enzyme ligation according to conventional methods, transformed into Escherichia coli DH5α, positive clones were selected, and DNA plasmids were prepared. The prepared D...
Embodiment 2
[0054] Mutation of the 516th amino acid arginine encoded on the HIV-1 gp160 gene to serine
[0055] Using the plasmid PAC-gp160 as a template, PCR reactions were performed with the following two pairs of primers, respectively.
[0056] Reaction 1:
[0057] Primer 1: 5' end primer (SEQ ID NO: 3) in Example 1
[0058] Primer 2: 5’GGA ACA AAG CTC CTA TTC CCA CTG C G C TTT TTTCTC TCT GC 3' (SEQ ID NO: 5) (note: the underlined G is T in the wild-type sequence)
[0059] Reaction 2:
[0060] Primer 3: 5'GTG GGA ATA GGA GCT TTG TTC CTT GG 3' (SEQ ID NO: 6).
[0061] Primer 4: 3' end primer (SEQ ID NO: 4) in Example 1.
[0062] The products obtained from PCR reaction 1 and reaction 2 were mixed as a template, and the primers (SEQID NO: 3 and 4) in Example 1 were used to carry out PCR reaction again. The resulting PCR product was digested with XbaI, and inserted into the plasmid pUC19 (purchased from New England Biolab Company) that had been digested with XbaI and blunted at the H...
Embodiment 3
[0066] Preparation of recombinant vaccinia virus with gp-160 gene
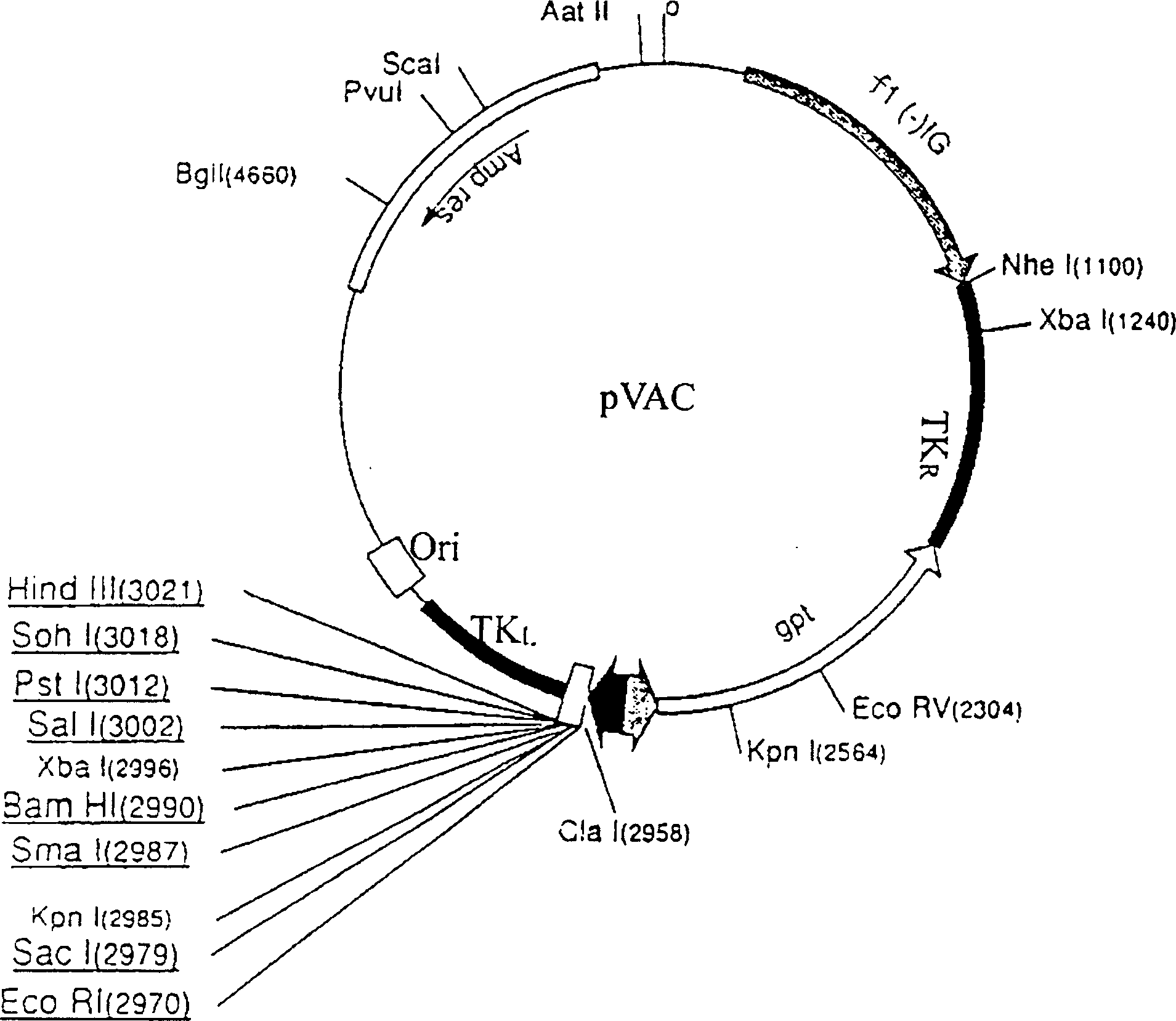
[0067] Digest PAC-gp160mu with endonuclease EcoRI, separate the gp-160mu DNA fragment, and insert it into the commonly used expression plasmid PVAC that has been digested by EcoRI (see the plasmid structure map image 3 )middle. The obtained PVAC plasmid carrying the gp-160mu gene was named PVAC-gp160mu.
[0068] Monkey kidney cell line BSC (ATCC CCL-26) was grown in complete DMEM medium to a density of 80%. BSC cells were infected with vaccinia virus New York strain (purchased from American Type Culture Collection, ATCC VR-1536) at a concentration of 0.1 PFU. The infected BSC cells were incubated in a 37°C carbon dioxide incubator for 2 hours. The PVAC-gp160mu plasmid DNA was transfected into BSC cells infected with vaccinia virus. The cells after DNA transfection were cultured in a carbon dioxide incubator at 37 degrees Celsius for 2 days, and then the cells were freeze-thawed and lysed, and then collect...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com