Eosinophil chemotactic factor
A technology of eosinophils and chemotaxis, applied in the field of genetic engineering, which can solve the problem of low-specificity migration of eosinophils and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
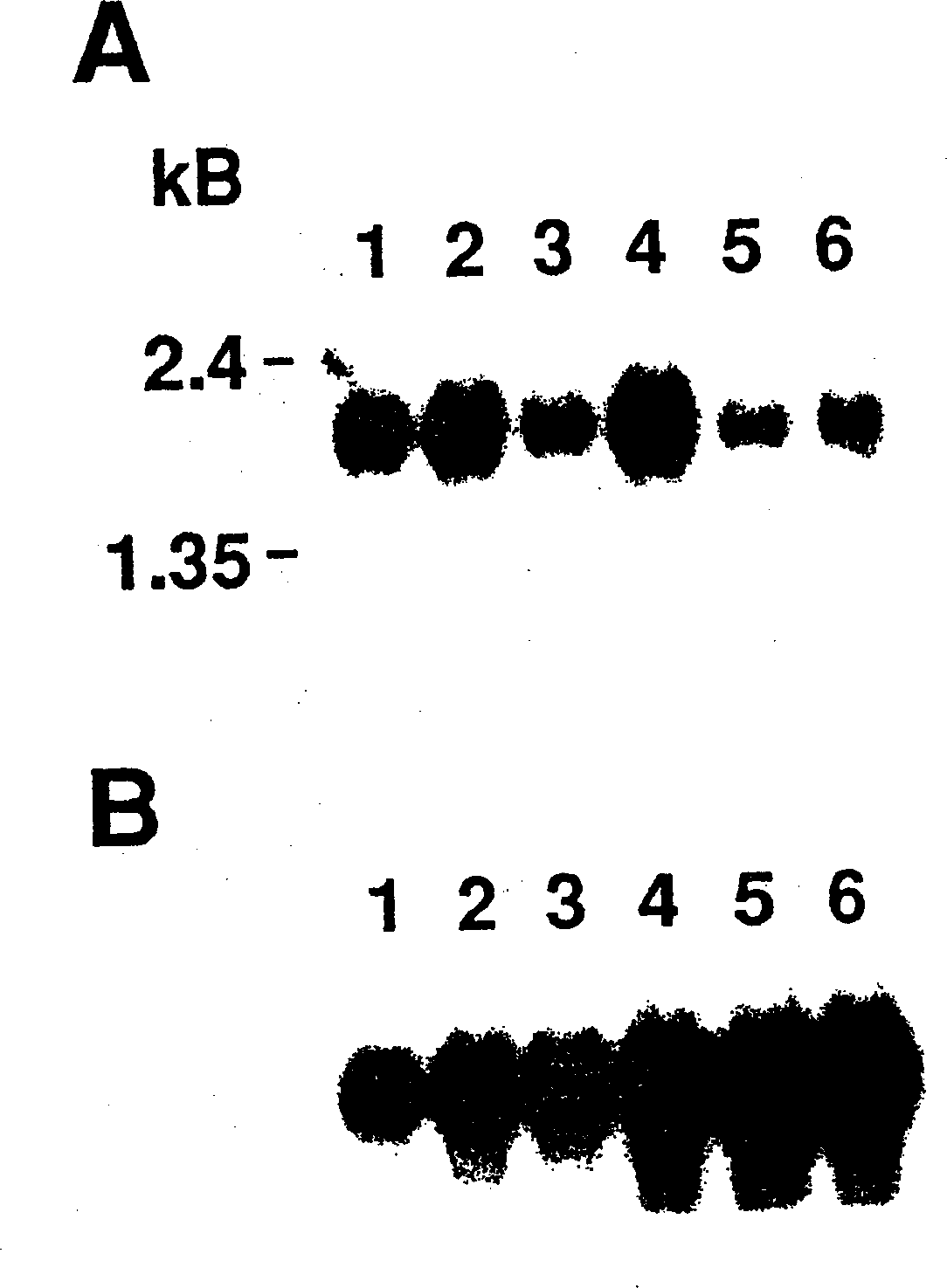
[0062] Embodiment 1: the preparation of the partial purification of ecalectin and the polyclonal antiserum containing ecalectin specific antibody
[0063] The cell line STO-2 obtained by transforming T cells of eosinophilic patients with human T-cell leukemia virus was used as a source material (Lymphokine Cytokine Research, 10, 481-486, 1991). FACS analysis confirmed that this cell line was derived from T cells, specifically, STO-2 expressed CD2, CD3, CD4, CD5, and CD8, but not CD16, CD19, or Leu7, markers for granulocytes / macrophages and B cells.
[0064] With the addition of 0.1% human serum albumin, 100 U / ml IL-2 (Tokushima Research Institute, Tokushima), 50 μM 2-mercaptoethanol, 100 μg / ml streptomycin, 100 U / ml penicillin and 5 μg / ml amphotericin B STO-2 cells were cultured in SF-02 serum-free medium for 72 hours, the culture supernatant was taken, and the pH was adjusted to 5 with 50 mM sodium acetate buffer. 5 liters of the conditioned medium thus obtained were passed ...
Embodiment 2
[0065] Example 2: Screening of cDNA library and positive clones prepared from T cell line STO-2
[0066] Use ZAP Express TM cDNA Gigapack Cloning Kit (Stratagene, La Jolla, California) was used to prepare the STO-2 cell cDNA expression library. Specifically, Fast Track RNA Isolation Kit (Invitrogen, San Diego, California) was used from 1 × 10 8 Collect about 5 μg poly(A) from each STO-2 cell + mRNA. Using Moloney mouse leukemia virus reverse transcriptase and 50-mer oligodeoxy dT primer (5'-GAGAGAGAGAGAGAGAGAGAACTAGTCTCGAGTTTTTTTTTTTTTTTTTT-3'; containing an XhoI restriction site: SEQ ID NO: 3), synthesize the first DNA strand. This DNA strand synthesis reaction is carried out in the presence of 5-methyl CIP to methylate the XhoI site in the cDNA. An EcoRI linker was ligated to the 5' end of the cDNA. Cut with restriction endonuclease XhoI (Stratagene), and carry out molecular sieve chromatography with 1ml Sephacryl S-500HR (Gibco) gel filtration column (balanced with 0....
Embodiment 3
[0072] Embodiment 3: Determination and analysis of nucleotide sequence
[0073] The DNAs of three independent clones obtained in Example 2 were assayed by the Sanger method (Proceedings of the National Academy of Sciences of the United States of America 74, 5463-5467), and it was found that they encoded the same protein. The DNA consensus sequence and its deduced amino acid sequence are shown in SEQ ID NO: 1 and SEQ ID NO: 2, respectively. The deduced open reading frame is believed to correspond to amino acids 60 to 1028 in SEQ ID NO: 1, which is predicted to encode a 36 KDa protein consisting of 323 amino acids. The present inventors named the cloned eotaxin as ecalectin. Nucleotides 1576 to 1581 in SEQ ID NO: 1 are a typical polyadenylation signal. In this clone, the 5' untranslated region has 59 nucleotides and the 3' untranslated region has 560 or more nucleotides. A repetitive sequence is often seen in the 3' untranslated region of short half-life transcripts (Cell, 46...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com