STAT2 gene deletion cell strain as well as preparation method and application thereof
A gene deletion and cell line technology, applied in the fields of molecular biology and genetics, can solve the problems of host cell tumorigenic risk, low efficiency of packaging virus, low efficiency of toxin production, etc., and achieve high knockout efficiency and strong reproducibility , the effect of reducing production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0092] The preparation method of the linear PUC-sgRNA plasmid is as follows: digest the PUC-sgRNA plasmid with BbsI.
[0093] The transformation and identification of sgRNA1 expression vectors and sgRNA2 expression vectors are carried out according to the following methods:
[0094] (1) Bacterial transformation
[0095] Preparation (~30 minutes in advance): take out the competent cells Trans-T1 (purchased from Quanshijin Company) stored in EP tubes from the -80°C refrigerator (100 μL per tube), and put them on ice to thaw (about 15 minutes) Finally, divide half (50 μL) of each tube into a new EP tube; turn on two water baths at 42 °C and 37 °C; take the resistant (Amp+) LB solid medium and put it in a 37 °C incubator to preheat.
[0096] Each tube of competent cells Trans-T1 (50 μL) was operated as follows: add 10 μL plasmid (i.e. sgRNA1 expression vector or sgRNA2 expression vector or reporter vector), place it on ice for 10 min, put it in a 42°C water bath, heat shock for 4...
Embodiment 1
[0102] This example is used to illustrate the cell line and its construction of STAT2 gene deletion
[0103] (1) STAT2 gene primer design
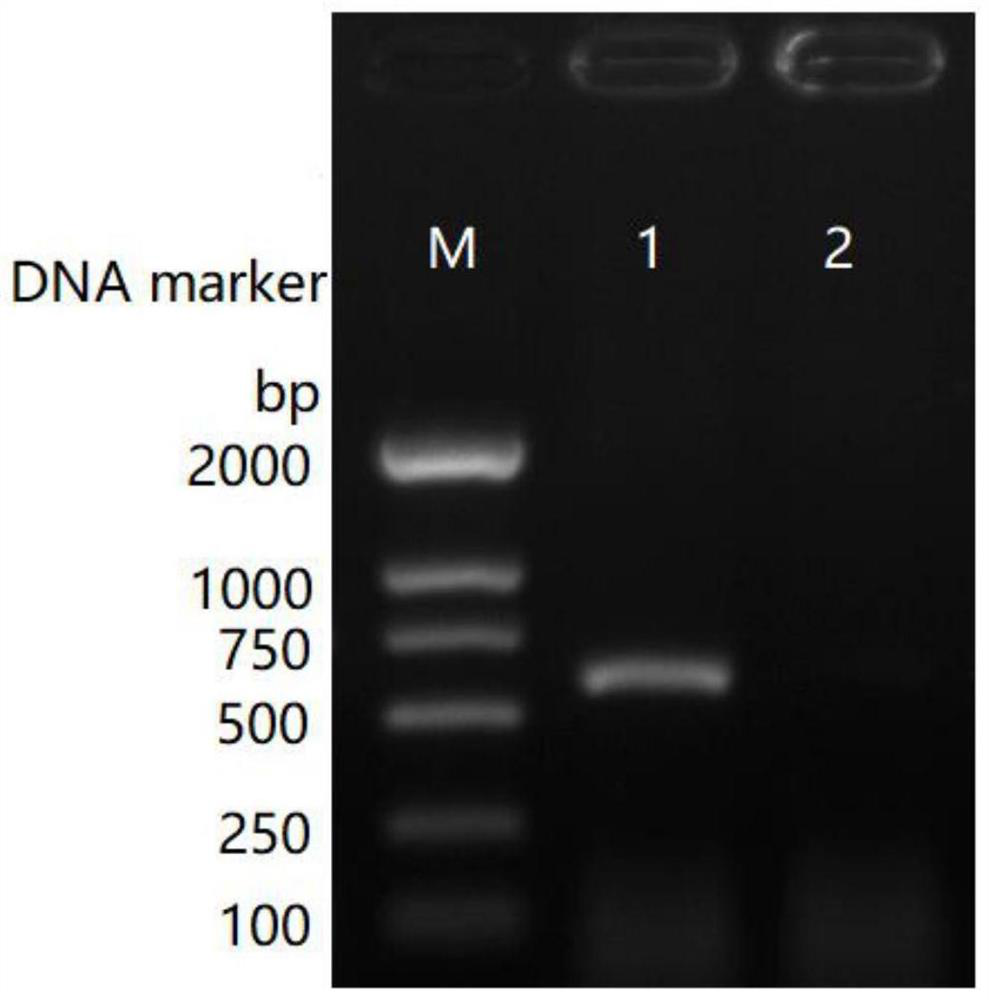
[0104] According to the STAT2 gene of HEK293 cells (STAT2 genome sequence NCBI ID: U18671.1), the primers in Table 1 were designed, and the primers were available (683bp) after PCR verification (such as figure 1 shown).
[0105] Table 1
[0106] Upstream primer: SEQ ID NO.:1(5'-3') TTGGGAACCCTCATCCTTCT Downstream primer: SEQ ID NO.:2(5'-3') CTTTGTCTTTTTCACCATAGC
[0107] (2) Construction of sgRNA1 expression vector and sgRNA2 expression vector
[0108] (a) sgRNA primer annealing (sgRNA primers annealling)
[0109] Two sgRNA targets were designed according to the STAT2 genome sequence, namely the sgRNA1 target sequence (SEQ ID NO.:3, GGAGGCTGTGCGAGTAAAGCTGG) and the sgRNA2 target sequence ((SEQ ID NO.:4, GATCAGCTGAACTATGAGTGTGG). A pair was synthesized according to the designed target sequence Primers (as shown i...
Embodiment 2
[0159] This example is used to illustrate the use of the HEK293 cell line with STAT2 gene deletion for virus packaging and effect verification.
[0160] (1) Lentiviral packaging
[0161] STAT2 gene-deleted HEK293 cells of the present invention were inoculated into 6-well plates for culturing as experimental group 1, and 293T cells (purchased from the Cell Bank of the Chinese Academy of Medical Sciences, GNHu17) were inoculated into 6-well plates for comparison as experimental group 2. Wild-type HEK293 cells were inoculated into 6-well plates for culture as experimental group 3. 24 hours before transfection, three groups of cells were plated with the same cell density (1x10 6 cells / well), when the cell density of the three groups reached about 80% confluence, the lentiviral plasmid packaging verification was carried out. The commercially available lentiviral packaging plasmid system (main plasmid PLVX-mCherry-C1, auxiliary plasmids PLP1, PLP2, PLP-VSVG, were purchased from Zh...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com