Neutralizing antibody for novel coronavirus SARS-CoV-2 and application of neutralizing antibody
A coronavirus, sars-cov-2 technology, applied in the field of biomedicine, can solve problems such as non-neutralization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0152] Example 1 - Screening of neutralizing antibodies
[0153] This embodiment is the screening of fully humanized scFv single chain antibody, and its steps include:
[0154] (1) Establishment of a fully human phage antibody library;
[0155] The peripheral blood of patients who had been infected with 2019-nCOV virus and recovered was collected, and the serum titer was determined. Select the sample with high titer to isolate lymphocytes, extract the total RNA of the cells, synthesize cDNA, use specific antibody heavy chain primers and light chain primers, amplify the variable region gene of the antibody, clone it into the phage display vector, and transfect into the host cell. Through NGS sequencing, the diversity and effectiveness of the antibody library were evaluated by quality control indicators such as library capacity, clone positive rate, heavy and light chain pairing, CDR3 distribution, and phage display rate.
[0156] (2) Phage library screening: use biotinylated...
Embodiment 2
[0161] Expression and purification of embodiment 2-scFv
[0162] In this embodiment, the 45 scFv single-chain antibodies screened in Example 1 were expressed and purified, which included the following steps:
[0163] Take the Escherichia coli TG1 bacterium liquid after shaking slightly, inoculate according to the ratio of 1:100, 37 ℃, 220rpm culture. When the OD is around 0.5, centrifuge at 8000rpm at room temperature for 5 minutes to obtain a bacterial liquid precipitate, induce with IPTG, culture at 30°C for about 12 hours, collect the expression supernatant for purification. The purification steps are as follows:
[0164] 1) Gently invert the bottle several times to mix the medium evenly;
[0165] 2) Draw a certain amount of medium and add it to the column, the column height is about 3-4 cm, and seal it when the medium settles to the bottom;
[0166] 3) Drain the storage solution, add 10mL equilibration buffer to equilibrate the chromatographic medium;
[0167] 4) Use a...
Embodiment 3-E
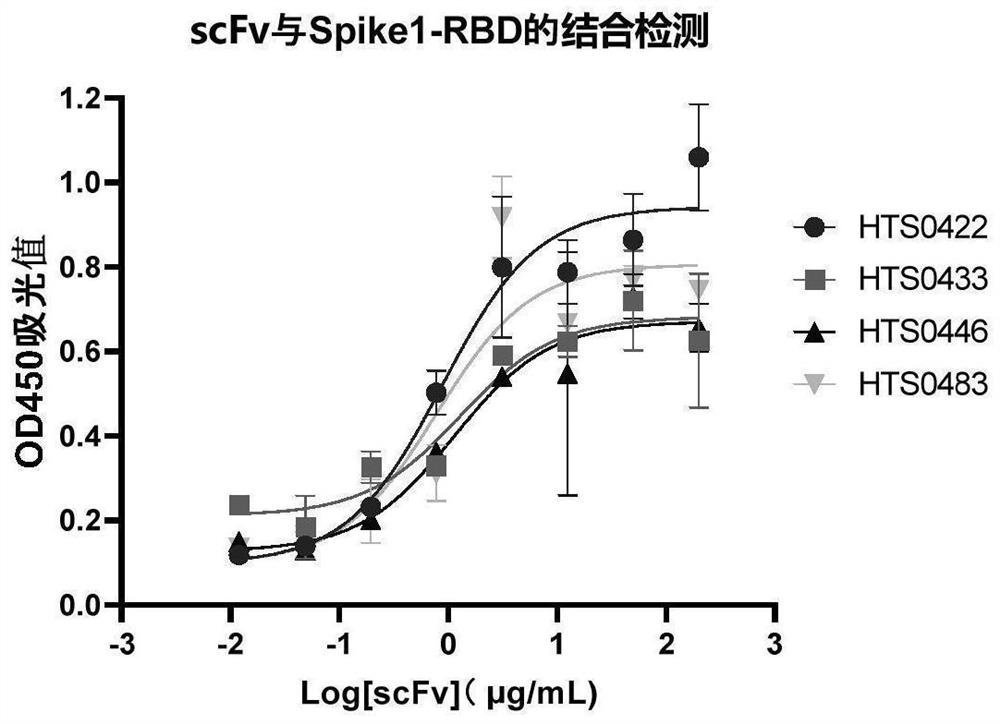
[0175] Embodiment 3-ELISA detects the binding activity of scFv
[0176] In this example, antibodies HTS0422, HTS0433, HTS0446 and HTS0483 were used as examples to detect the binding ability of purified scFv to Spike1-RBD antigen by ELISA. Specific steps are as follows:
[0177] 1) 384-well plate was coated with antigen, and 25 μL of 1 μg / mL Spike1-RBD antigen was added to each well, sealed, and coated overnight at 4°C;
[0178] 2) Discard the supernatant, wash 3 times with 100 μL Wash Buffer per well;
[0179] 3) 25 μL of blocking solution per well, block at room temperature for 1 hour, discard the liquid from the blocked ELISA plate, and wash 3 times with 100 μL of WashBuffer per well;
[0180] 4) Dilute the scFv antibody, the highest concentration is 200 μg / ml, four-fold serial dilution, a total of 8 points;
[0181] 5) Add 25 μL / well of the diluted antibody into appropriate wells and incubate at room temperature for 2 hours;
[0182] 6) Discard the supernatant, dry the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com