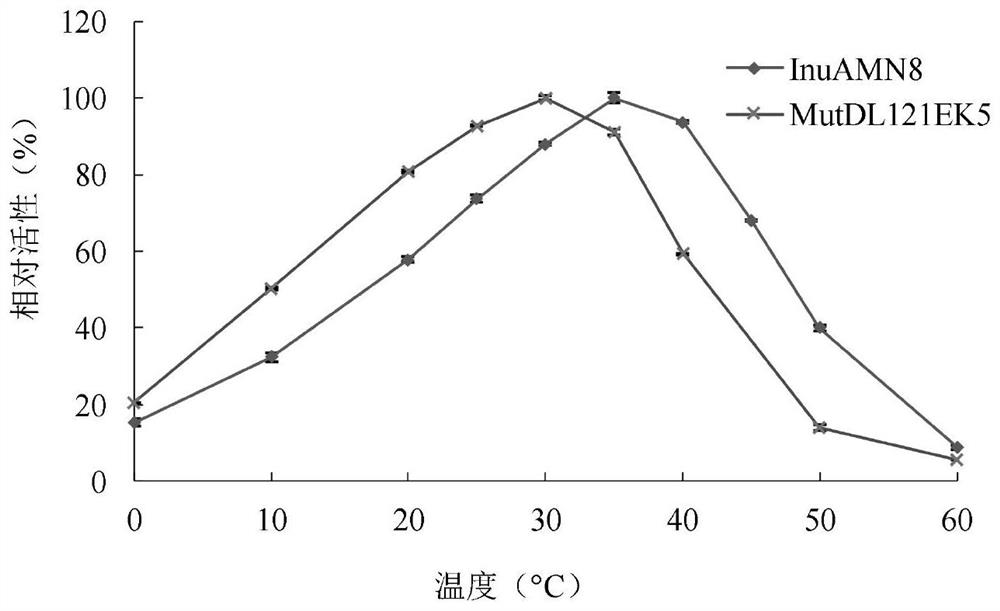
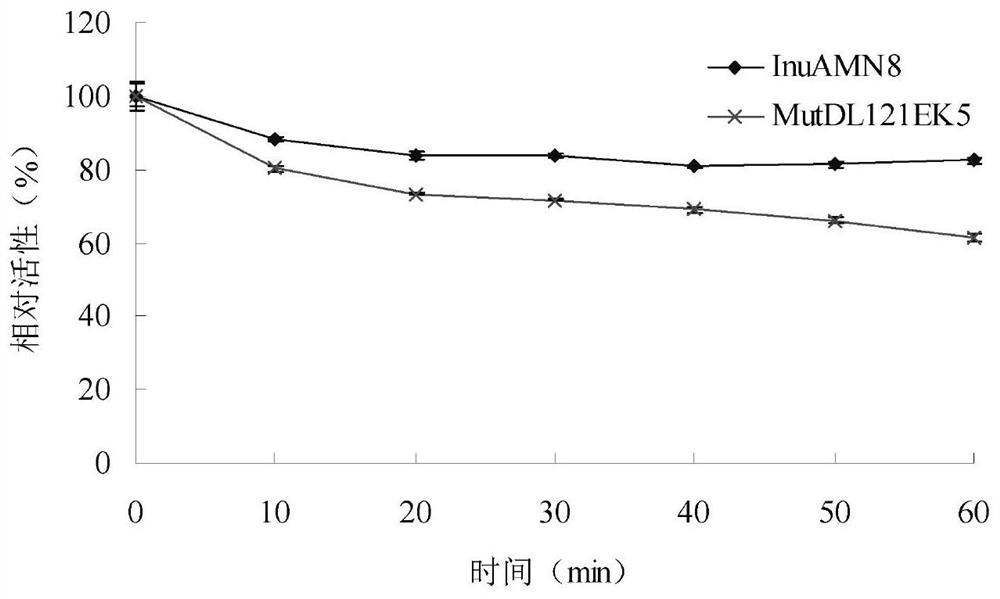
Low-temperature exoinulinase mutant MutDL121EK5 with improved low-temperature adaptability and application thereof
An exo-inulinase and mutant technology, which is applied in the field of genetic engineering and can solve the problems of easy degradation of enzymes and easy exposure of protease cleavage sites.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Construction and Transformation of Embodiment 1 Wild Enzyme InuAMN8 Expression Vector
[0035] 1) Extraction of Arthrobacter genomic DNA: Centrifuge the bacterial liquid cultured for 2 days to obtain the bacterial cells, add 1 mL of lysozyme, treat at 37°C for 60 minutes, and then follow the instructions of the bacterial genomic DNA extraction kit (Tiangen Biochemical Technology (Beijing) Co., Ltd.) Genomic DNA of Arthrobacter was extracted and stored at -20°C for later use.
[0036] 2) According to the exo-inulinase nucleotide sequence JQ863111 (SEQID NO.4) recorded in GenBank, primers 5'ATGAATTCATTGACGACGGC 3' and 5'TCAACGGCCGACGACGTCGA 3' were designed, and the Arthrobacter genomic DNA was used as a template for PCR amplification. The reaction parameters are: denaturation at 95°C for 5 min; then denaturation at 95°C for 30 sec, annealing at 58°C for 30 sec, extension at 72°C for 1 min and 30 sec, and after 30 cycles, keep at 72°C for 5 min. The PCR results obtained ...
Embodiment 2
[0039] Construction and transformation of embodiment 2 mutant enzyme MutDL121EK5 expression vector
[0040] 1) Design primers 5'TGAAGAAGACCGAAAGCCGGGCCGGCAGGCGCAG 3' and 5'GCTTTCGGTCTTTCCTACTGTAGGCACTGGTGTAAATGGC 3', and use the plasmid pEasy-E1-inuAMN8 as a template for PCR amplification. The PCR reaction parameters are: denaturation at 95°C for 30 sec; denaturation at 95°C for 15 sec, annealing at 70°C for 15 sec , extend at 72°C for 3min 30sec, and after 30 cycles, keep at 72°C for 5min. As a result of PCR, the recombinant expression linearized plasmid pEasy-E1-mutDL121EK5 containing mutDL121EK5 was obtained. mutDL121EK5 and pEasy-E1-mutDL121EK5 can also be obtained by gene synthesis.
[0041] 2) Add 1 μL of DpnI enzyme to 50 μL of the PCR product of the linearized plasmid pEasy-E1-mutDL121EK5, and digest it at 37° C. for 1 h.
[0042] 3) Use Mut II Fast Mutagenesis Kit, place the digested product in (2) at 37°C for 30min.
[0043] 4) The ligation product in (3) was tr...
Embodiment 3
[0044] Embodiment 3 Preparation of recombinant wild enzyme InuAMN8 and mutant enzyme MutDL121EK5
[0045] The recombinant strains BL21(DE3) / inuAMN8 and BL21(DE3) / mutDL121EK5 were inoculated in LB (containing 100 μg mL -1 Amp) medium, shake rapidly at 37°C for 16h.
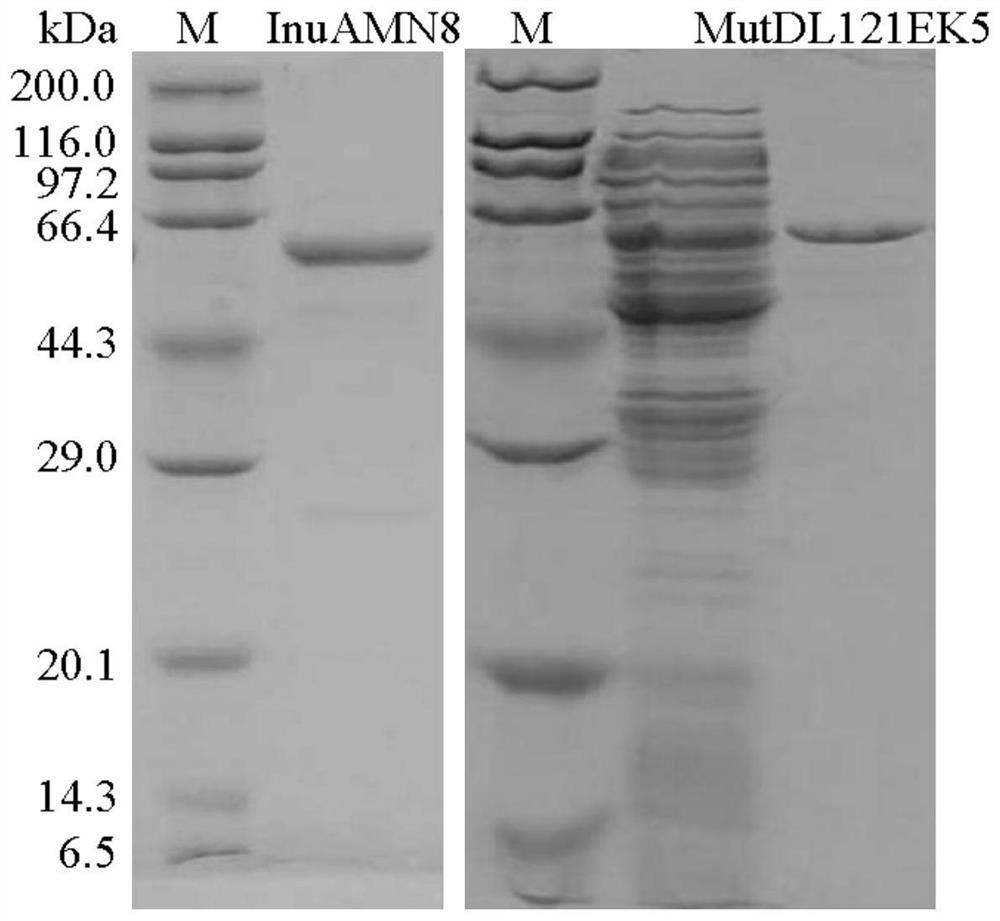
[0046] Then the activated bacterial solution was inoculated into fresh LB (containing 100 μg mL -1 Amp) culture solution, after rapid shaking culture for about 2-3 hours (OD600 reaches 0.6-1.0), add IPTG with a final concentration of 0.7mM for induction, and continue shaking culture at 20°C for about 20 hours. Centrifuge at 12000rpm for 5min to collect the bacteria. After suspending the cells with an appropriate amount of McIlvaine buffer with pH=7.0, the cells were ultrasonically disrupted in a low-temperature water bath. After the crude enzyme solution concentrated in the cells was centrifuged at 13,000 rpm for 10 min, the supernatant was aspirated and the target protein was affinity-purified with Nickel-NTAAg...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com