DNA template for modifying primary cells by gene editing and fixed-point insertion method
A gene editing and DNA sequence technology, applied in the field of gene-directed insertion, can solve the problems of complex, uneconomical, and dangerous preparation of double-stranded linear DNA, and achieve great clinical application potential and commercial value, good safety and effectiveness. performance and cost savings
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
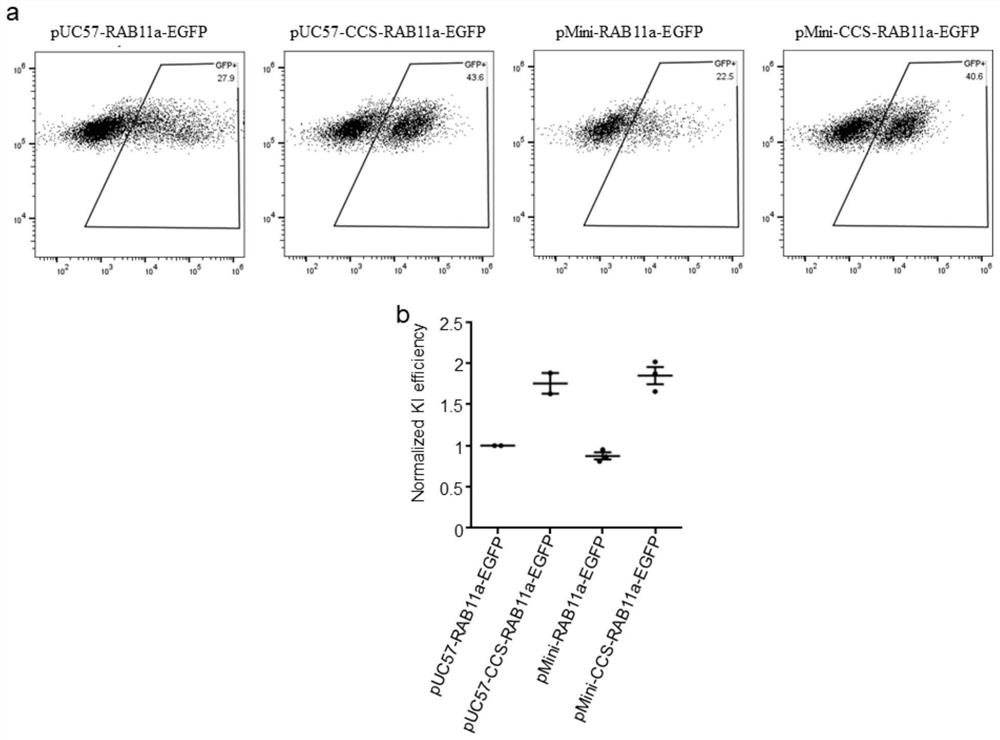
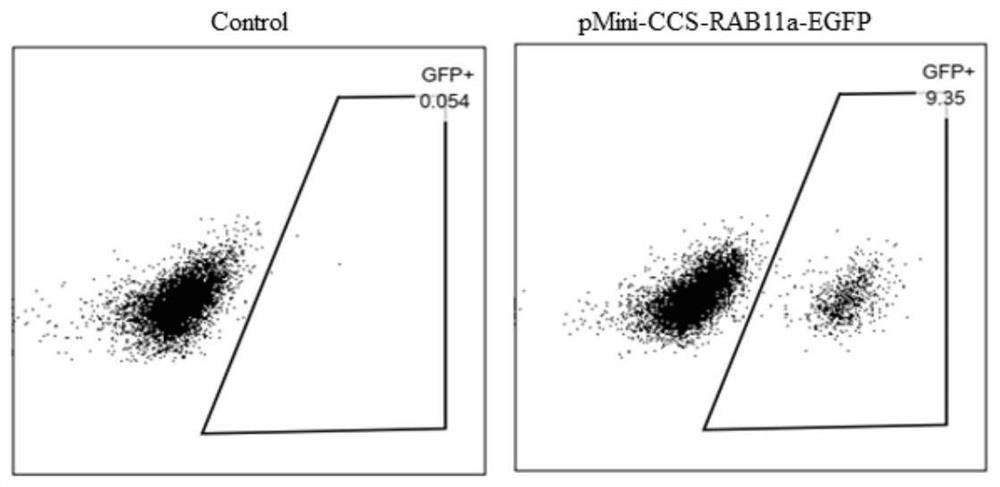
[0047] 1. Construction of EGFP gene and RAB11a homology arm sequence on pUC57 or transformed pMini plasmid vector 1. Transformation of pUC57 plasmid vector sequence
[0048] The pUC57 plasmid vector sequence is 2.7kb in size and mainly includes three parts: the lactose operon sequence (LacZ), the ampicillin resistance gene and its promoter region (AMP), and the prokaryotic origin of replication (ORI). On the basis of existing research, this example shrinks the pUC57 plasmid vector, removes the lactose operon sequence (LacZ) sequence by molecular cloning, and PCR clones the ORI region forward primer sequence as shown in SEQ ID NO: 1, reverse See SEQ ID NO: 2 for the primer sequence, see SEQ ID NO: 3 for the forward primer sequence of the cloned AMP region, and see SEQ ID NO: 4 for the reverse primer sequence. The pUC57 truncated mutant plasmid was constructed by homologous recombination and named pMini. See SEQ ID NO:5 for the sequence of pMini.
[0049] 2. Construction of don...
Embodiment 2
[0056] 1. Synthesize the guide RNA sequence of the first exon of RAB11a gene.
[0057] Two, the SpCas9 (2NLS and 4NLS) used in the present embodiment and the LbCpf1 or AsCpf1 protein purification used in Example 4 are mainly divided into the following two steps:
[0058] 1. Construct the Cas9 / Cpf1 plasmid expression vector, express the Cas9 / Cpf1 protein with the Escherichia coli plasmid expression vector system, and amplify the Cas9 / Cpf1 gene from the pX330 plasmid vector by PCR. The forward primer sequence is shown in SEQ ID NO: 14, and the reverse For the sequence of the forward primer, see SEQ ID NO: 15, for the sequence of the forward primer of LbCpf1, see SEQ ID NO: 16, for the sequence of the reverse primer, see SEQ ID NO: 17, for the sequence of the forward primer of AsCpf1, see SEQ ID NO: 18, for the sequence of the reverse primer See SEQ ID NO:19. Recover after PCR amplification, connect the recovered SpCas9, LbCpf1, and AsCpf1 fragments to the KS expression plasmid ...
Embodiment 3
[0067] 1. Synthesis of CAR gene and homology arm gene
[0068] Taking CD19-CAR targeting CD19 currently used in clinical treatment of leukemia as an example, the amino acid sequence of the CAR gene is shown in SEQ ID NO: 23, mainly including the ScFv that recognizes the specific tumor antigen CD19, and the extracellular segment of CD28 , the transmembrane region and the intracellular segment, and the intracellular segment of CD3ζ. The gene sequence of the left homology arm is shown in SEQ ID NO:24, and the gene sequence of the right homology arm is shown in SEQ ID NO:25. The CAR gene and homologous arm gene fragments were synthesized by PCR, the forward primer was synthesized by PCR as shown in SEQ ID NO:26, and the reverse primer was as shown in SEQ ID NO:27.
[0069] 2. Construction of CAR gene and homology arm sequence on pUC57 / pMini / pMiniZ plasmid vector
[0070] 1. Clone the homology arm sequences on both sides of the target gene CAR and TCRα into the pUC57 / pMini / pMiniZ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com