Molecular ruler hairpin structure and method for measuring spatial scale accuracy of single-molecule magnetic tweezers by using same
A technology of spatial scale and hairpin structure, which is applied in the fields of biochemical equipment and methods, determination/inspection of microorganisms, instruments, etc., can solve the problems of the accuracy of the spatial scale of single-molecule magnetic tweezers, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
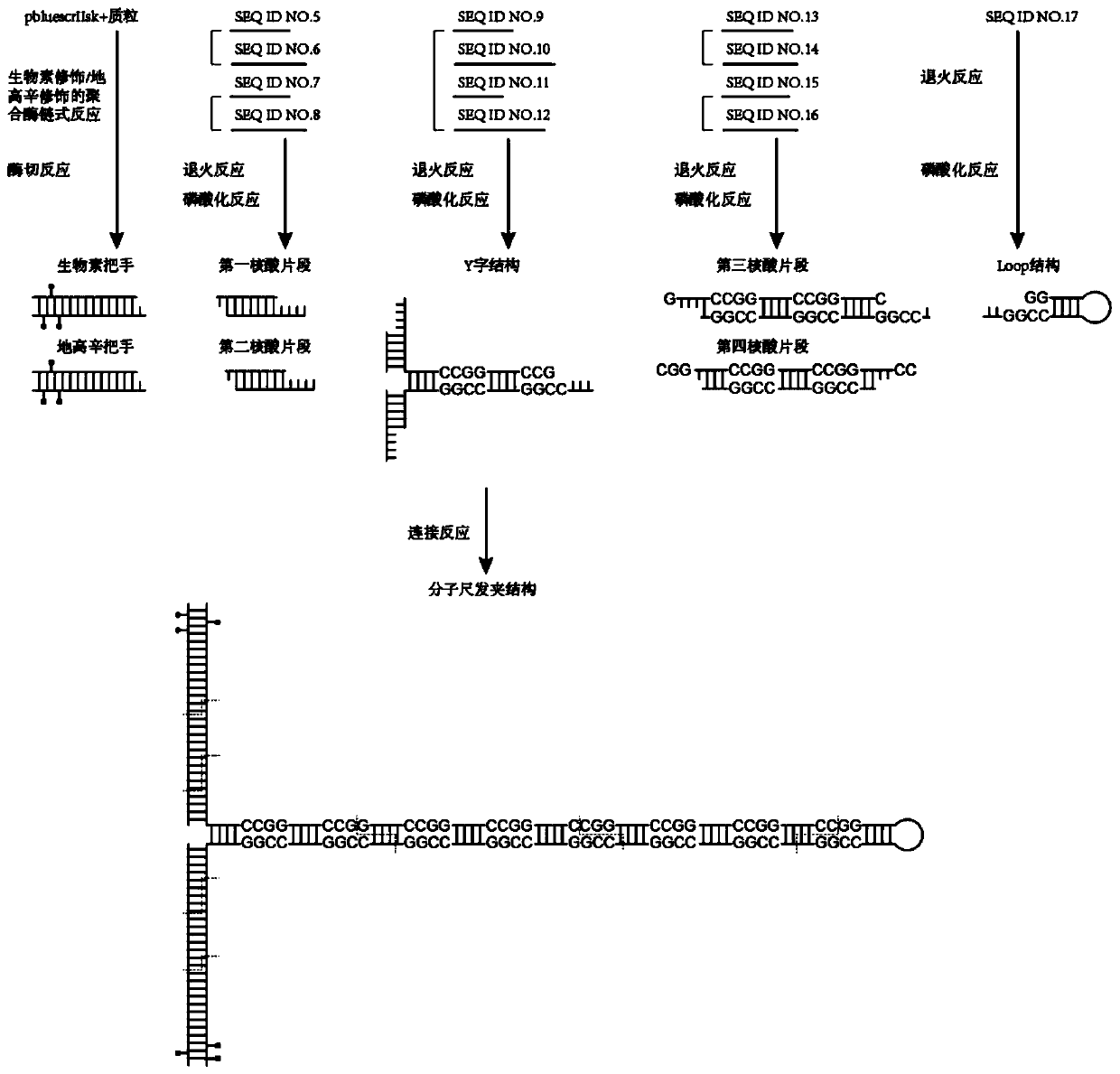
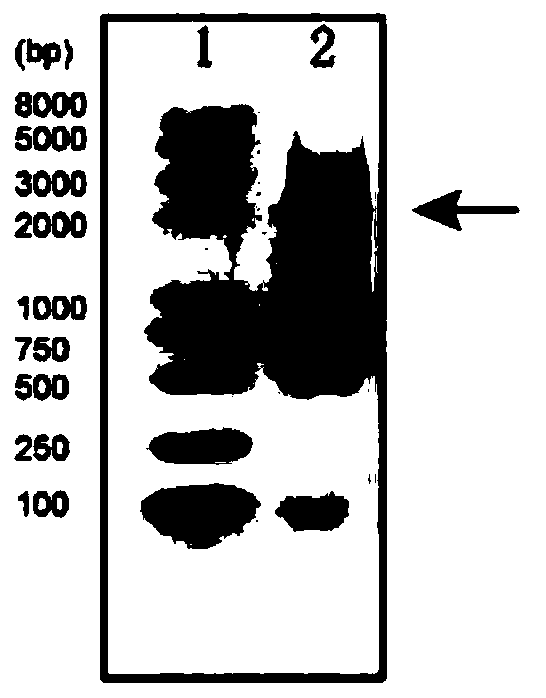
[0082] Embodiment 1 Construction of molecular ruler hairpin structure
[0083] 1 Preparation of biotin handle and digoxin handle
[0084] 1.1 Polymerase chain reaction of biotin modification / digoxigenin modification
[0085] Perform biotin-modified / digoxigenin-modified PCR on the pbluescrIISK+ plasmid by adding:
[0086] 1 μl of primers upstream of the biotin / digoxigenin handle at a concentration of 10 μM, respectively shown in SEQ ID NO.1 and SEQ ID NO.3, (primers purchased from Jinweizhi);
[0087] 1 μl of primers downstream of the biotin / digoxigenin handle at a concentration of 10 μM, as shown in SEQ ID NO.2 or SEQ ID NO.4, respectively (primers were purchased from Jinweizhi Company);
[0088] 1 μl 10mM dATP (Catalog No.: 4026Q, Baori Medical Biotechnology Company);
[0089] 1 μl 10mM dGTP (Catalog No.: 4027Q, Baori Medical Biotechnology Company);
[0090] 1 μl 10mM dCTP (Catalog No.: 4028Q, Baori Medical Biotechnology Company);
[0091] 0.9 μl 10mM dTTP (Catalog No.: ...
Embodiment 2
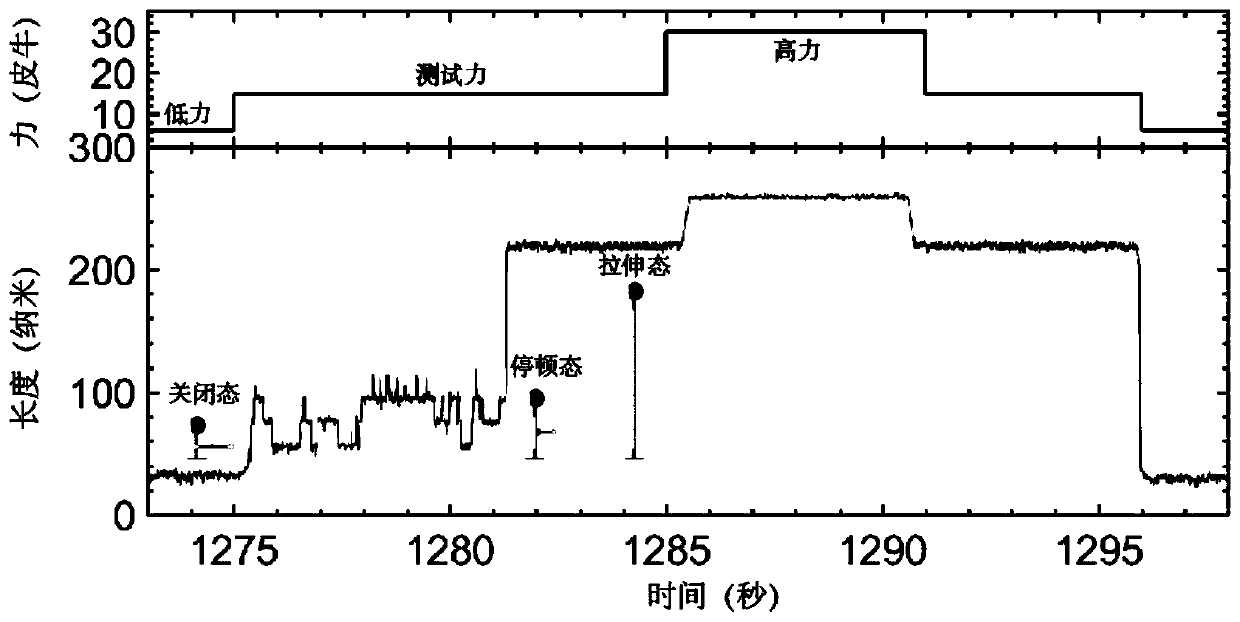
[0169] Example 2 Measure and calculate the measured conversion coefficient of nano / base by applying external force with single-molecule magnetic tweezers
[0170] In this embodiment, the magnetic tweezers experiment uses a magnetic tweezers device produced by Tianjin Guangying Micro-Nano Technology Co., Ltd. The molecular ruler hairpin structure can be fixed on the glass surface of the working cell through the interaction between digoxin-digoxigenin antibody, and the biotin-streptavidin affinity binding with superparamagnetic beads (M270, item number: 65305, Invitrogen Corporation) connected.
[0171] The preparation process of the working pool is as follows: take two clean coverslips of 60mm×24mm, and use a TC-169 multi-function grinding and engraving machine to punch two holes on one piece as the water inlet and water outlet, and evenly smear 3μm polystyrene beads on one piece As a magnetic bead reference, a double-layer sealing film with a reaction channel cut in advance i...
Embodiment 3
[0174] Example 3 Calculate the theoretical conversion coefficient of nanometers / bases according to the theoretical worm model
[0175] The DNA length can be calculated by the theoretical worm model, as shown in Equation 1, where F is the force, x is the length, Lp is the persistent length, Lc is the profile length, and K B is the Boltzmann constant, and T is the temperature.
[0176]
[0177]According to Bosco, A. Under the working conditions described in the article Elastic properties and secondary structure formation of single-stranded DNA at monovalent and divalent salt conditions (PMID: 24225314), Lp is selected as 0.87 nanometers, and Lc is 0.69 nanometers / base, according to This calculated theoretical conversion factor is 0.48 nm / base.
[0178] The deviation between the measured conversion coefficient and the theoretical conversion coefficient in this embodiment is 4%. Therefore, the spatial scale accuracy of the single-molecule magnetic tweezers used in the instrume...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com