Fluorescent probe for quantitatively detecting acidic or basic amino acids on basis of carbon quantum dot fluorescence quenching or enhancement method and preparation method for fluorescent probe
A carbon quantum dot and fluorescence quenching technology, applied in the field of fluorescent probes, can solve the problems of slow growth, mental retardation, stunting, etc., and achieve the effects of low price, simple preparation method, and easy procurement.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
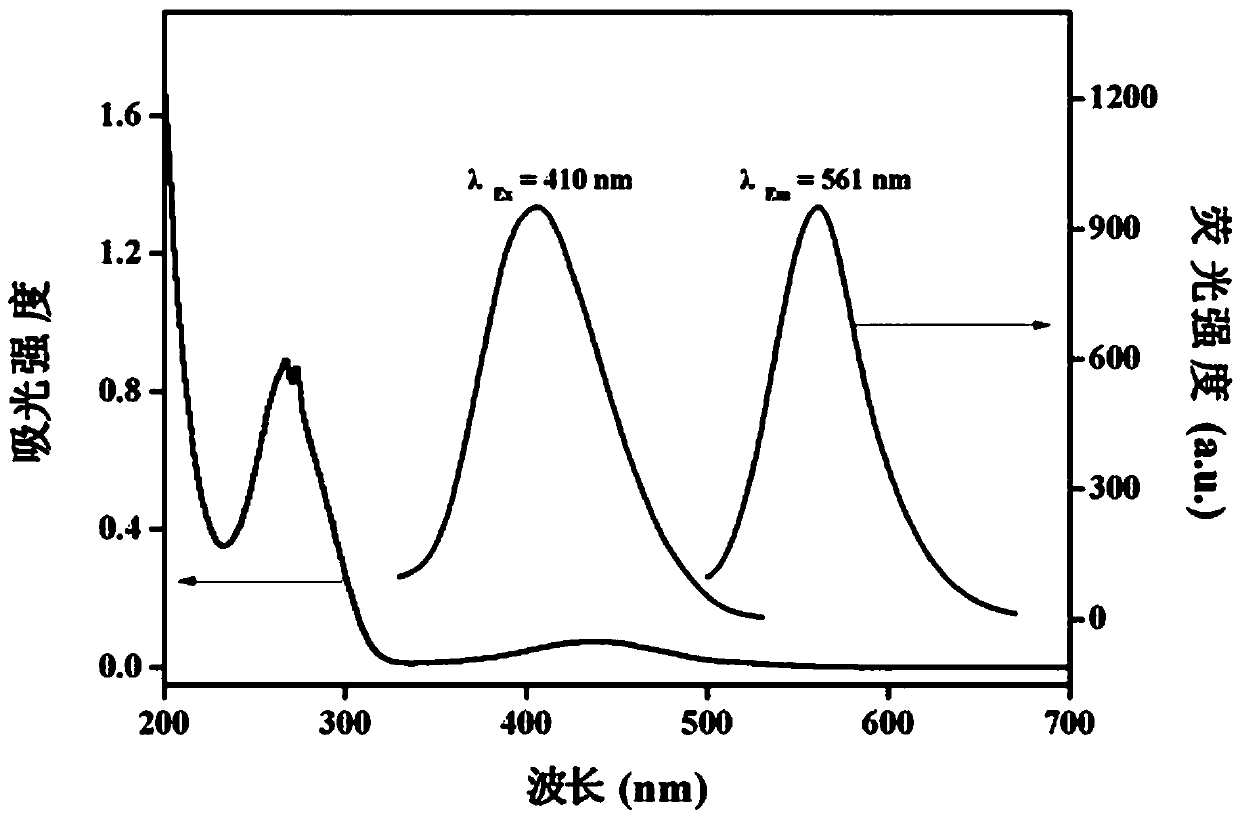
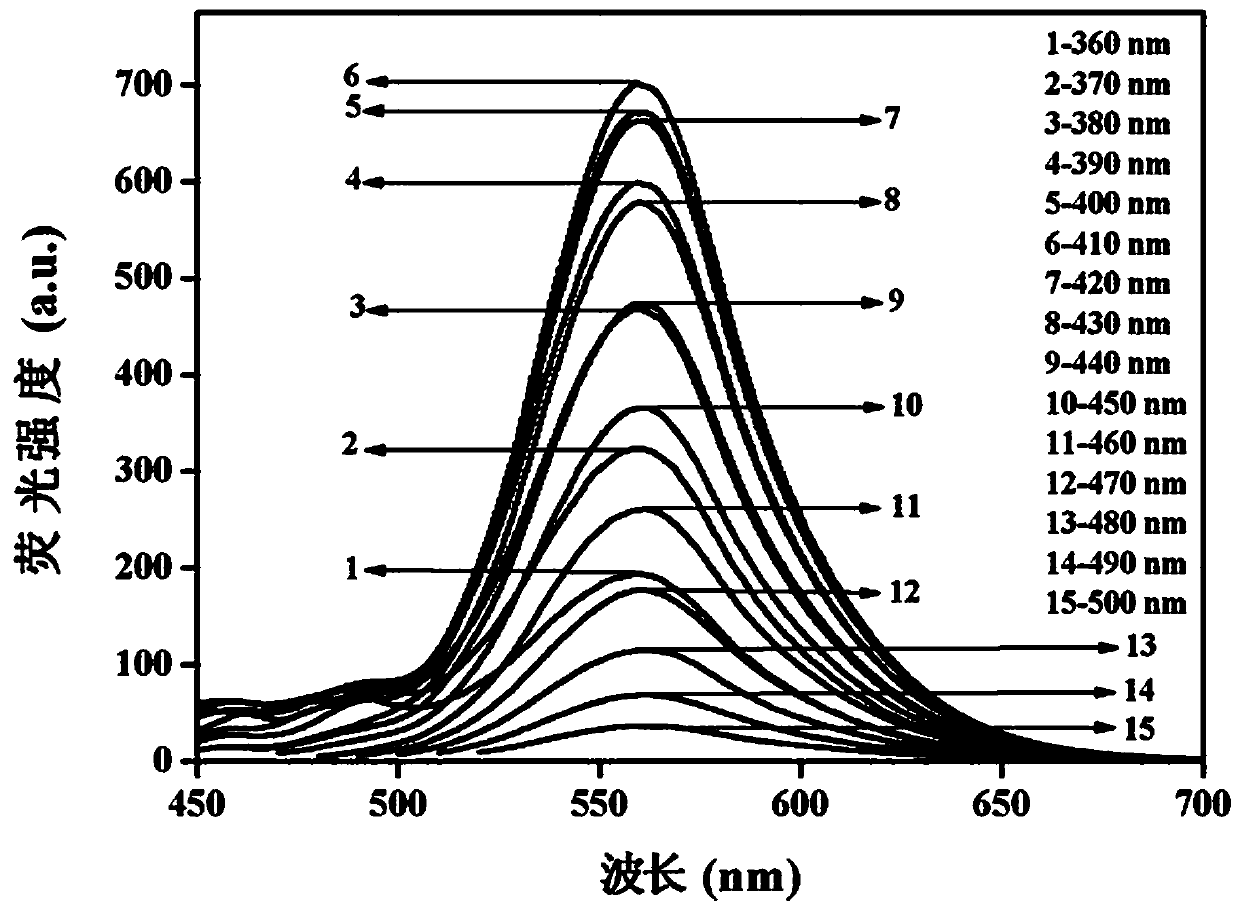
[0040] Example 1: Preparation and Characterization of C-dots
[0041] Step 1: Accurately weigh 0.05 g of o-phenylenediamine and 0.05 g of p-aminobenzoic acid, add 10 mL of absolute ethanol to them, dissolve them fully by ultrasonication (200W, 10 min), and transfer them to 100 mL of polytetrafluoroethylene lining , assemble the reactor and put it in the oven, at 180 o C for 12 h.
[0042] Step 2: After the reactor is cooled to room temperature, the solution in the reactor is rotatably evaporated to remove absolute ethanol. Add 10 mL of secondary water, sonicate (200W, 10 min) to dissolve the sticky matter and transfer it to a centrifuge tube, centrifuge the solution at 8000 rpm for 10 minutes, transfer the supernatant to a beaker, and freeze-dry to obtain C-dots solid powder.
[0043] Step 3: Weigh 0.1 g of C-dots solid powder into a beaker, add 10 mL of secondary water to it, and ultrasonically (200W, 10 min) dissolve it fully to obtain a C-dots stock solution with a conce...
Embodiment 2
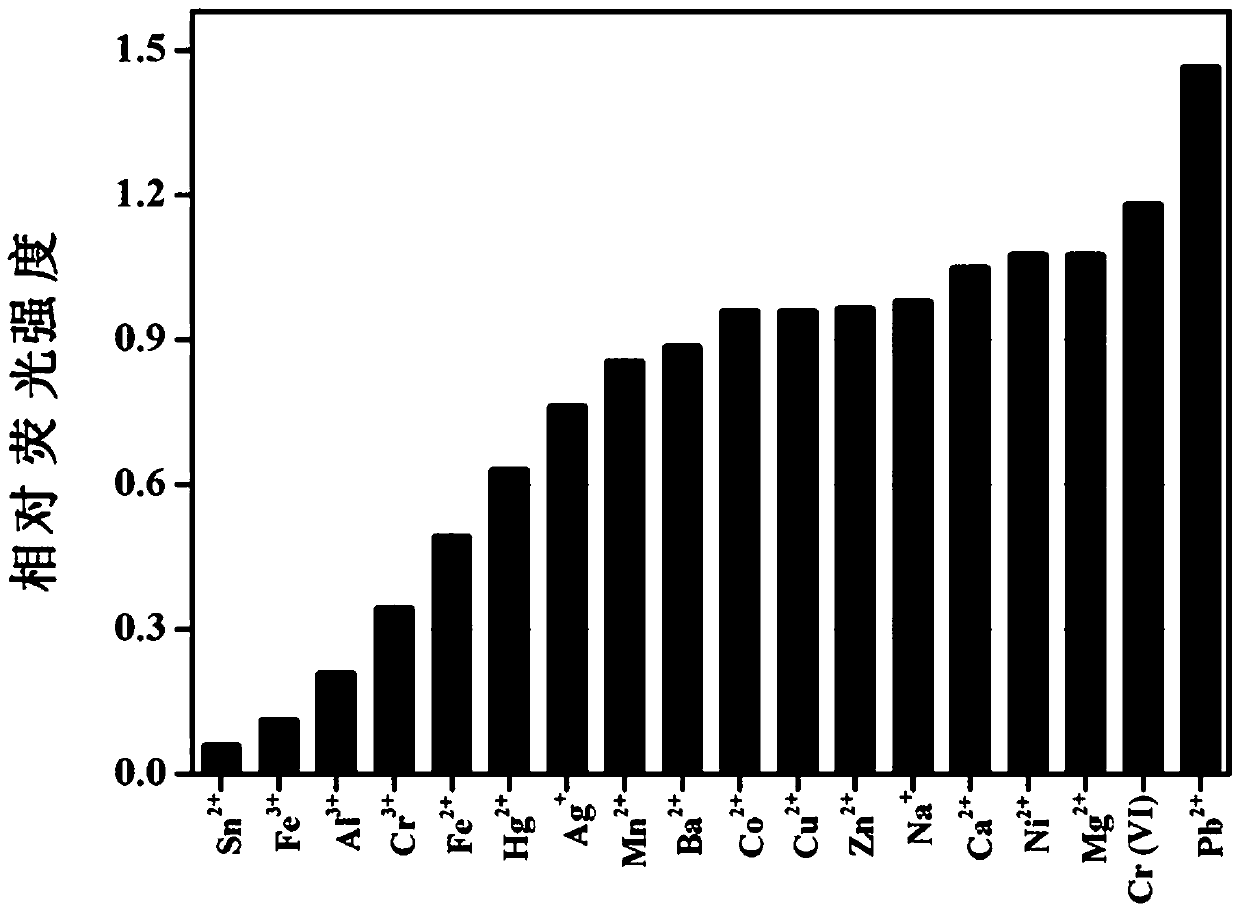
[0046] Example 2: Response of different substances to C-dots
[0047] Step 1, accurately weigh a certain mass of metal ions (Sn 2+ , Fe 3+ , Al 3+ , Cr 3+ , Fe 2+ , Hg 2+ , Ag + , Mn 2+ , Ba 2+ , Co 2+ , Cu 2+ , Zn 2+ , Na + , Ca 2+ , Ni 2+ , Mg 2+ , Cr(VI), Pb 2+ ) compound, add 10 mL of secondary water, ultrasonically (200W, 5 min) fully dissolve, and prepare a metal ion stock solution with a molar concentration of 0.1 mol / L.
[0048] Step 2: Accurately weigh a certain mass of amino acids (aspartic acid, glutamic acid, tryptophan, phenylalanine, methionine, cysteine, proline, isoleucine, serine, valine amino acid, threonine, leucine, glycine, alanine, homocysteine, tyrosine, asparagine, glutamine, histidine, arginine, lysine), adding 10 mL of secondary water, ultrasonically (200W, 5 min) to fully dissolve, and prepare an amino acid stock solution with a molar concentration of 0.01 mol / L.
[0049] Step 3, accurately weigh a certain mass of anion (C 2 o 4 ...
Embodiment 3
[0056] Example 3: C-dots for titration of acidic and basic amino acids
[0057] Step 1, add 7 µL of C-dots (10 mg / mL) stock solution to 2 mL of secondary water, at this time the concentration of C-dots is 0.0349 mg / mL (2007 µL), test and record the fluorescence intensity of C-dots f 0 .
[0058] Step 2, add aspartic acid stock solution drop by drop to the solution in step 1, test and record the fluorescence intensity F of C-dots, draw the fluorescence spectrum diagram of the whole titration process, see the experimental results Figure 9 . Calculation (F 0 -F) / F 0 And take it as the ordinate, the concentration of aspartic acid as the abscissa, utilize Origin fitting to obtain the linear equation, (F 0 -F) / F 0 = 0.0080c (Asp) +0.0699, R 2 =0.9989, the linear range is 10.94-29.80 μmol / L, and the lowest detection limit is 0.1925 umol / L. See the experimental results Figure 10 .
[0059] Step 3, add glutamic acid stock solution drop by drop to the solution in step 1, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com