Novel lipase as well as preparation and application thereof
A lipase and mutant technology, applied in the direction of application, enzyme, hydrolase, etc., to achieve high-efficiency expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
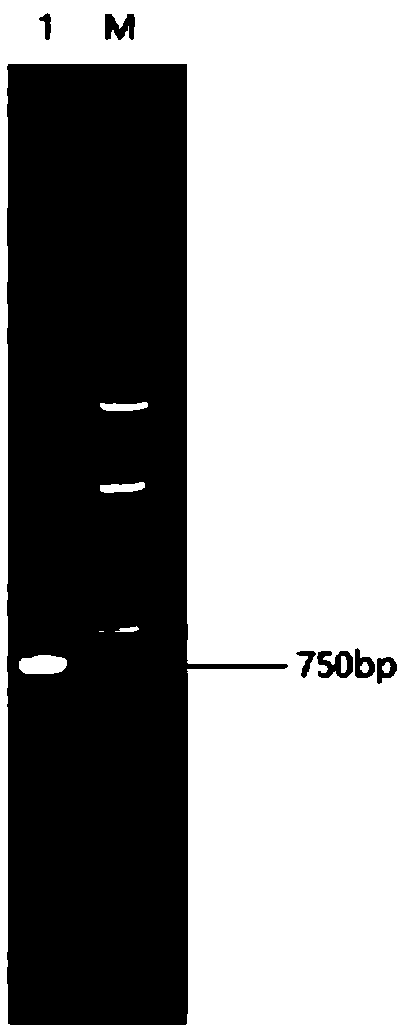
[0061] Embodiment 1: the acquisition of wild-type lipase gene
[0062] 1. The wild-type lipase gene is from Penicillium cyclopium CICC 41049 strain, and its total RNA is extracted.
[0063] (1) Strain activation: draw 100 μL of preserved Penicillium arcuci spore liquid from the glycerol tube, evenly spread it in the PDA eggplant bottle, and incubate at a constant temperature of 28°C for 5 days;
[0064] (2) Transfer: Wash the spores in the eggplant bottle with sterile water, centrifuge at 12000r / min for 1min, wash repeatedly, and finally transfer to 50mL PDA liquid medium, place in a shaker, 28°C, 200r / min min, cultivated for 2 days;
[0065] (3) Collect the bacteria: filter the bacteria with double-layer sterilized gauze, rinse with sterile water, wring out, place the bacteria in a mortar, add liquid nitrogen and grind to powder;
[0066] (4) Add Trizol: Take a small amount of the ground powder and put them into EP tubes, add 1mL Trizol reagent into each tube, shake the nes...
Embodiment 2
[0094] Embodiment 2: Obtaining of lipase mutant N157F
[0095] 1. Ligate the codon-optimized wild-type lipase gene of Escherichia coli to the pET-22b(+) vector.
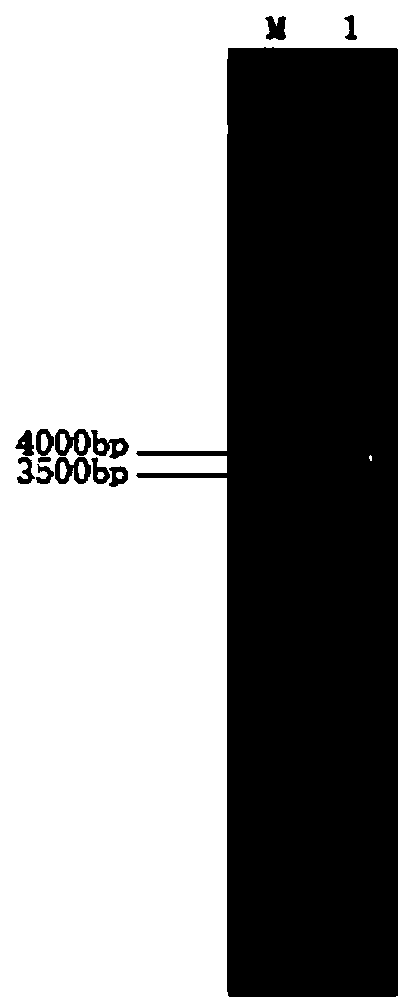
[0096] The purified pcl was ligated with the pET-22b(+) vector, and then the recombinant plasmid was transformed into Escherichia coli DH 5α, and the codon-optimized wild-type fat of Escherichia coli was successfully verified by double digestion with BamH I and Hind III The enzyme gene has been cloned into the pET-22b(+) vector to construct the recombinant plasmid pET-pcl.
[0097] 2. Error-prone PCR: Using the recombinant plasmid pET-pcl constructed above as a template, the reaction system is as follows:
[0098] wxya 2 o
21μL
Recombinant plasmid pET-pcl (5ng / μL)
1μL
Upstream primer P1 (10μmol / L)
2μL
Downstream primer P2 (10μmol / L)
2μL
0.5μL
10×Taq buffer
5μL
dATP (10mmol / L)
1μL
dGTP (10mmol / L)
1μL
...
Embodiment 3
[0112] Embodiment 3: the construction of the lipase recombinant bacterium that Bacillus subtilis alkali resistance improves
[0113] 1. Construction of expression vector pBSA43
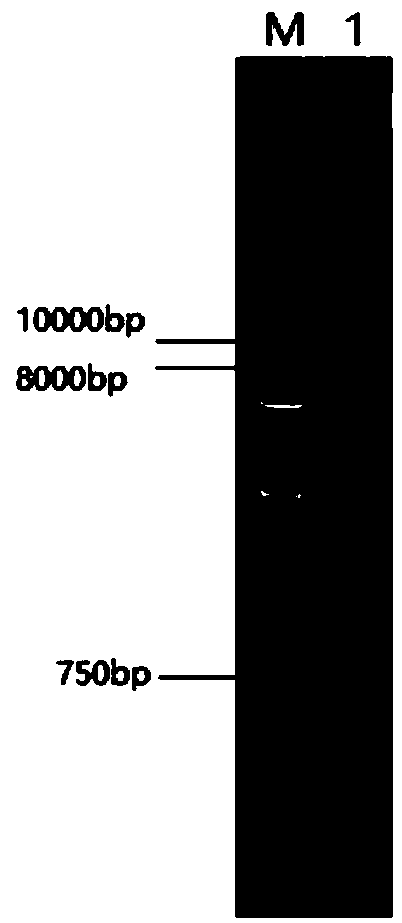
[0114] Using the Escherichia coli-Bacillus subtilis shuttle cloning vector pBE2 as the backbone, clone into a strong Bacillus constitutive promoter P43 (SEQ ID No.10) and fructan sucrase that can directly secrete the recombinant protein into the medium The signal sequence sacB (SEQ ID No.11) was obtained from the expression vector pBSA43. it comes with amp r and Kana r Gene, ampicillin resistance can be used as a selection marker in Escherichia coli, and kanamycin resistance can be used as a selection marker in Bacillus subtilis and Bacillus licheniformis.
[0115] 2. Construction of lipase expression plasmid pBSA43-Bsmpcl with improved alkali resistance
[0116] The codon-optimized lipase mutant gene Bsmpcl (SEQ ID No.6) of Bacillus subtilis and the expression vector pBSA43 of Bacillus subtilis w...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Extend | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com