Genetic engineering subunit vaccine for porcine circovirus as well as preparation method and application of genetic engineering subunit vaccine
A porcine circovirus and subunit vaccine technology, applied in vaccines, antiviral agents, veterinary vaccines, etc., to achieve the effects of reduced virus content, simple preparation process, and high purity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
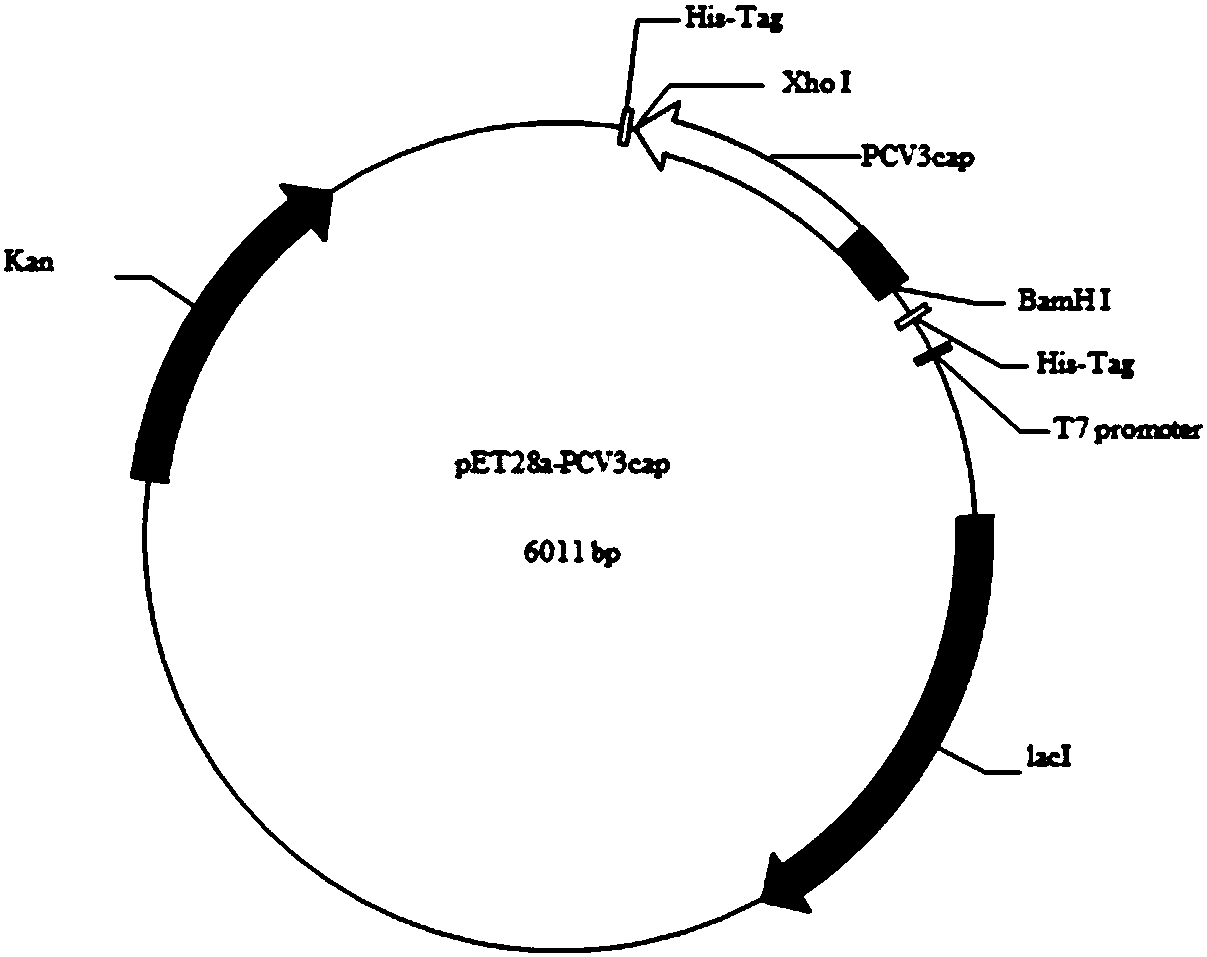
[0042] Example 1 Expression of recombinant Cap protein by E. coli
[0043] 1. Gene Amplification
[0044] Amplify Cap gene with PCV3-China / GD2016 virus strain (PCV3-China / GD2016 genome information has been disclosed in Genomecharacterization of a porcine circovirus type 3 in South China, Changxu Song, Transbound Emerg Dis. 2017 Mar 13. in the paper) as template, The reaction system is shown in Table 1, and the reaction program is shown in Table 2.
[0045] Table 1 Reaction system for Cap gene amplification
[0046]
[0047] Among them, the upstream primer is: 5'-GGCggatccATGAGACACAGACCTATATTC-3', the downstream primer is: 5'-GGGctcgagGAGAACTGACTTGTAACGAAT-3', the lowercase letters indicate the restriction sites BamHI and XhoI respectively.
[0048] Table 2 PCR reaction program for Cap gene
[0049]
[0050] (3) Gel electrophoresis of PCR amplification products
[0051] Referring to the method of "Molecular Cloning Experiment Guide", prepare a 1.2% agarose gel, take 5 μ...
Embodiment 2
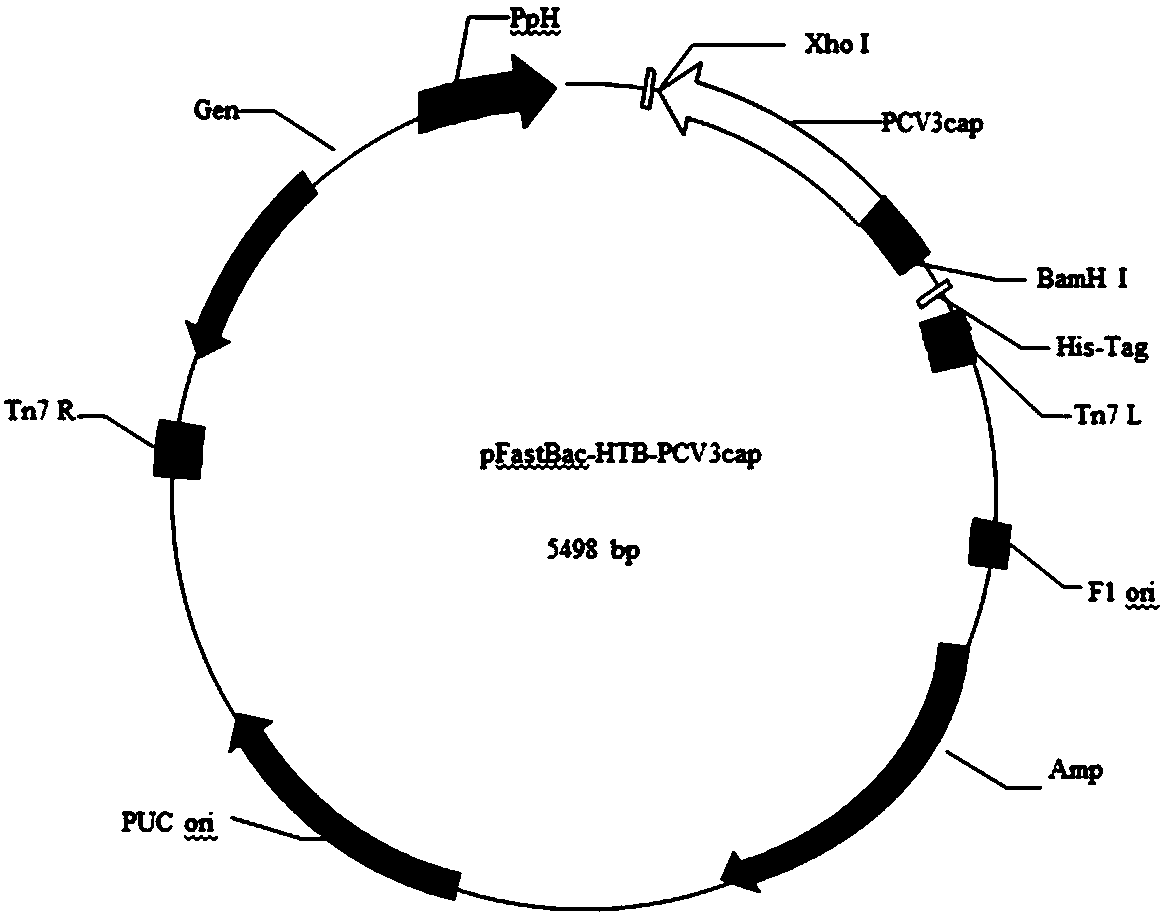
[0120] Example 2 Baculovirus expresses recombinant Cap protein
[0121] 1. Gene Amplification
[0122] Same as Example 1.
[0123] 2. Cap target fragment gel recovery
[0124] Same as Example 1.
[0125] 3. Cap fragment and pFastBac-HTB double digestion
[0126] The obtained Cap gene fragment and pFastBac-HTB plasmid were digested with BamHI and XhoI, respectively, and the digestion systems are shown in Table 6 and Table 7.
[0127] Table 6 Double digestion reaction system of pFastBac-HTB plasmid
[0128]
[0129] Table 7 Double enzyme digestion reaction system of Cap gene fragment
[0130]
[0131] Incubate in a metal bath at 37°C for 20 min. After the reaction is completed, the digestion product is analyzed by agarose gel electrophoresis according to the molecular cloning manual, and the target fragment is recovered. For the recovery method, refer to the instructions of the gel recovery kit (Omega company).
[0132] 4. Cap fragment and pFastBac-HTB ligation
[0...
Embodiment 3
[0189] Embodiment 3 Vaccine immune effect experiment
[0190] Fifteen healthy susceptible piglets about 3 weeks old, without PCV3 antigen and antibody, without PRRSV, PEDV, PRV, Staphylococcus suis and other major swine pathogens, were selected and randomly divided into 3 groups. The piglets were injected after mixing equal volumes of immunogen and adjuvant according to Table 10 before injection. 3 weeks after immunization, serum was collected to determine Cap-specific antibody titers; 1 week after blood collection, challenge was performed. The challenge strain was PCV3-China / GD2016, and the titer was 10^ 6 TCID50, 1 mL per head, intranasally, 14 days after challenge, lymph nodes were collected, and the content of PCV3 in the tissue was determined by fluorescence quantitative PCR.
[0191] Table 10 Immune effect experiments
[0192]
[0193] The specific operation of the fluorescent quantitative PCR method is as follows: Design quantitative primers for the Cap protein ge...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com