DNA (deoxyribonucleic acid) molecular markers for identifying early bolting of angelica sinensis and application of DNA molecular markers
A technology of DNA molecule and Angelica sinensis, applied in the field of identification of early shoots of Angelica sinensis, can solve the problems affecting the yield and quality of Angelica sinensis, and the identification method of early shoots of Angelica sinensis has not been reported, and achieves good stability and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] DNA molecular marker sequence screening method of Angelica early sprouts:
[0025] 1. Total DNA Extraction from Angelica Genome
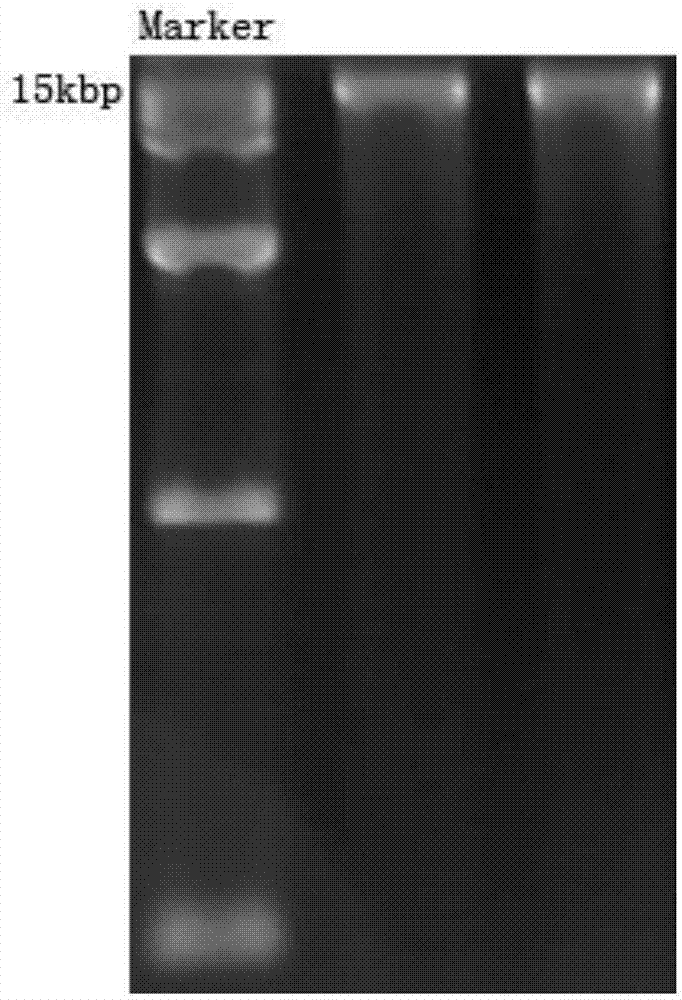
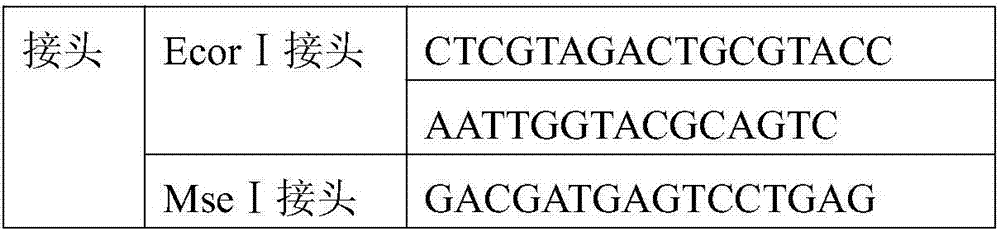
[0026] Plant genomic DNA was extracted using the modified CTAB method. Weigh 1 g of Angelica sinensis leaves, grind them into powder with liquid nitrogen, collect the powder in a centrifuge tube, add 4 mL of CTAB extract preheated to 65 °C, and incubate at 65 °C for 1 h. Add an equal volume of chloroform / isoamyl alcohol (24:1) for extraction, invert to mix well, centrifuge at 5500rpm / min for 10min, take the supernatant, and repeat the extraction 2-3 times. Aspirate the supernatant and add 2 / 3 volume of pre-cooled isopropanol, and let stand at -20°C for 2h. Centrifuge at 7500g for 10min, dissolve the precipitate with 400μLTE, add 4μL of pre-boiled RNaseA, and incubate at 37°C for 30min. Then extract with an equal volume of chloroform / isoamyl alcohol (24:1) for 1-2 times, take the supernatant and add an equal volume of isopropanol to precipi...
Embodiment 2
[0092] Primers were designed for the molecular marker ZT-2, and PCR amplification was performed using Angelica sinensis DNA as a template. The primer sequence is, Primer-F:
[0093] CCGTAGGCTGATCCTTTCCCT, Primer-R: CCATGAACCAAGTGCCTTCAC. The PCR amplification reaction was carried out with the genome DNA of early sprouts and normal angelica as templates. Reaction conditions, denaturation at 94°C for 3min; denaturation at 94°C for 30s, annealing at 58°C for 30s, extension at 72°C for 1min (40 rings). Electrophoresis was performed on the PCR products, and the labeling rate of Angelica zaponicae was 75%. Example 3
Embodiment 3
[0094] Primers were designed for the molecular marker ZT-3, and PCR amplification was performed using Angelica sinensis DNA as a template. The primer sequence is, Primer-F:
[0095] GAGTACCTAGGCGGAGTACA, Primer-R:ACACCCAATAACCATCACCC. The PCR amplification reaction was carried out with the genome DNA of early sprouts and normal angelica as templates. Reaction conditions, denaturation at 94°C for 3min; denaturation at 94°C for 30s, annealing at 58°C for 30s, extension at 72°C for 1min (40 rings). Electrophoresis was performed on the PCR products, and the labeling rate of Angelica zaponicae was 70%. Example 4
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com