Immunogenic polypeptide surface layer-expressing bifidobacterium
An immunogenic peptide, bifidobacteria technology, applied in the direction of peptide sources, bacteria, bacterial peptides, etc., can solve the problems of large side effects and long treatment period
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0147] (preparation of transformed bifidobacteria)
[0148] Hereinafter, an example of the preparation procedure of a transformed bifidobacterium in which an immunogenic polypeptide of interest is expressed and presented as a fusion protein on the surface of bifidobacteria will be described.
[0149] 1. Acquisition of genes
[0150] The gene encoding GLBP and the gene encoding the immunogenic polypeptide of interest can be obtained based on known gene sequence or amino acid sequence information, respectively. For example, using genomic DNA or cDNA prepared from any bifidobacterium as a template, using a primer pair prepared based on the sequence information of the GLBP structural gene of the bifidobacterium, amplified by polymerase chain reaction (PCR) to obtain . Generally, since multiple codons exist for one amino acid, a gene may have a base sequence different from a known base sequence or a base sequence based on a known amino acid sequence.
[0151] For example, the ge...
Embodiment 1
[0201] (Example 1: Design of HCVNS3 polypeptide gene for bifidobacterium surface layer expression)
[0202] design figure 2 The two amino acid sequences shown are based on the NS3 linker site (1196-1215 positions) of the HCV-1b type polypeptide and the β-α-β structural domain (1216-1350 positions) on the upstream side, which will be derived from the NS3 Antigenic peptides are linked to the N-terminus and C-terminus of the base domain.
[0203] Amino acid sequence 1( figure 2 >1: SEQ ID NO: 23) The N-terminal side of the basic domain is connected to a region containing QSFLATCINGVCWTVYHGAG (1067-1086, CD8 epitope 1: SEQ ID NO: 4), and the C-terminal side is connected to EIPFYGKAI (1372 ~1380 position, CD8 epi position 4: sequence number 7), KLSALGVNA (position 1406~1414, CD8 epi position 6: sequence number 9) and VATDALMTGYTGDFDSVIDC (position 1435~1454, CD8 epi position 7: sequence number 10) area.
[0204] Amino acid sequence 2( figure 2 >2: SEQ ID NO: 24) A region co...
Embodiment 2
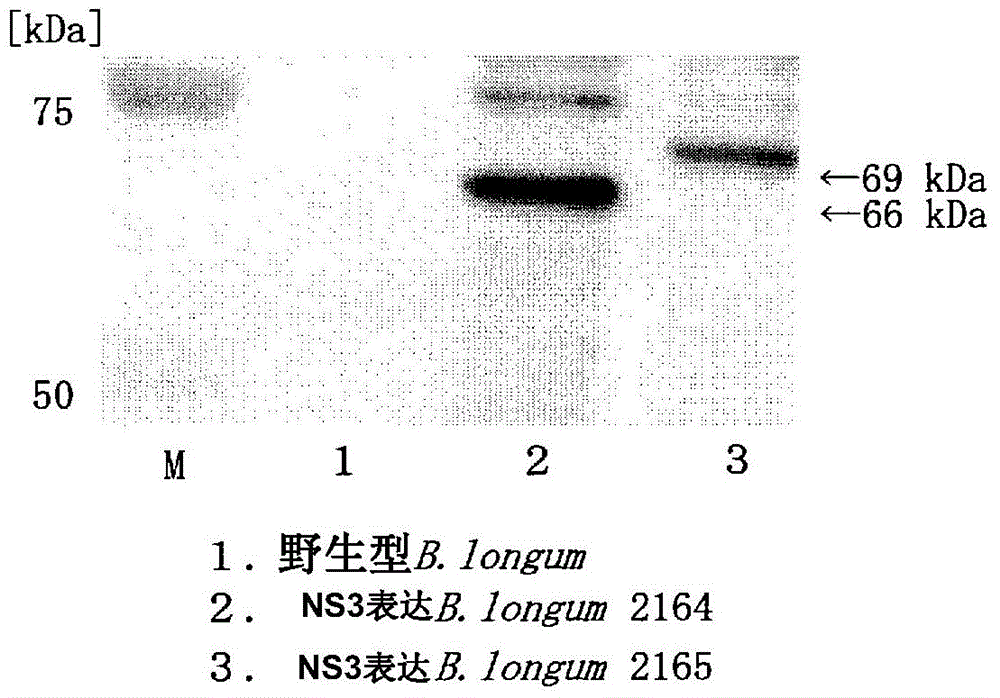
[0206] (Example 2: Preparation of NS3 Protein Surface Expression Transformed Bifidobacteria)
[0207] Based on the amino acid sequences 1 and 2 (sequence numbers 23 and 24, respectively) designed in Example 1, a gene sequence conforming to the codon usage frequency (http: / / www.kazusa.or.jp / codon / ) of bifidobacteria was designed 1 and 2 (sequence numbers 25 and 27 respectively; corresponding amino acid sequences are represented by sequence numbers 26 and 28), based on these gene sequence information, full-length synthesis of respective gene fragments (the former is also referred to as "long form", the The latter is called "short form"). The full-length synthesis of the gene fragments was entrusted to GenScript Corporation. The obtained gene fragment was treated with XhoI and SalI, and inserted into recombinant plasmid pJT101 treated with XhoI and SalI (Patent Document 4 and Non-Patent Document 4). Plasmid pJT101 contains the GLBP gene derived from Bifidobacterium longum JCM12...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com