Double-lipase cell surface co-display engineering bacterium, and construction method and application thereof
A technology of cell surface and lipase, which is applied in the field of double lipase cell surface co-display engineering bacteria and its construction, can solve the problems of high cost, loss of activity, complicated extraction separation and purification process, etc., and achieve high catalytic efficiency and catalytic reaction short time effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Saccharomyces cerevisiae S.cerevisiaeEBY100 genome extraction, the steps are as follows:
[0035] (1) S. cerevisiae EBY100 was purchased from Invitrogen. First, activate the strain on the YPD plate, pick a single colony in the YPD shake flask, and culture overnight at 30°C.
[0036] (2) The bacterial cells in the logarithmic growth phase were centrifuged at 5000 rpm / min for 5 minutes, the bacterial cells were washed once with sterile distilled water, and the supernatant was removed.
[0037] (3) Add 5 mL of DNA breaking buffer (100 mmol / LTris-HCl, pH 8.0, 10 mmol / LEDTA, 1% SDS), mix well, and incubate at 65° C. for 1 h.
[0038] (4) Centrifuge at 10,000 rpm for 15 minutes, and take the supernatant.
[0039] (5) Add phenol chloroform isoamyl alcohol (25:24:1) to extract twice, and centrifuge at 5000rpm for 7min each time.
[0040] (6) Take the upper layer extract, add 6 μL RNaseA (10 mg / mL), and precipitate with 3 times ethanol overnight (add 0.3 mol / L sodium acetate,...
Embodiment 2
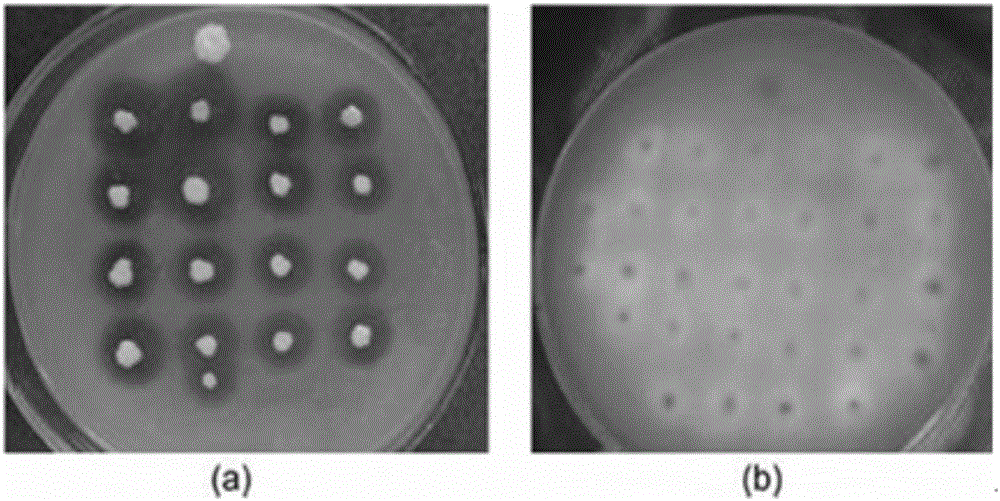
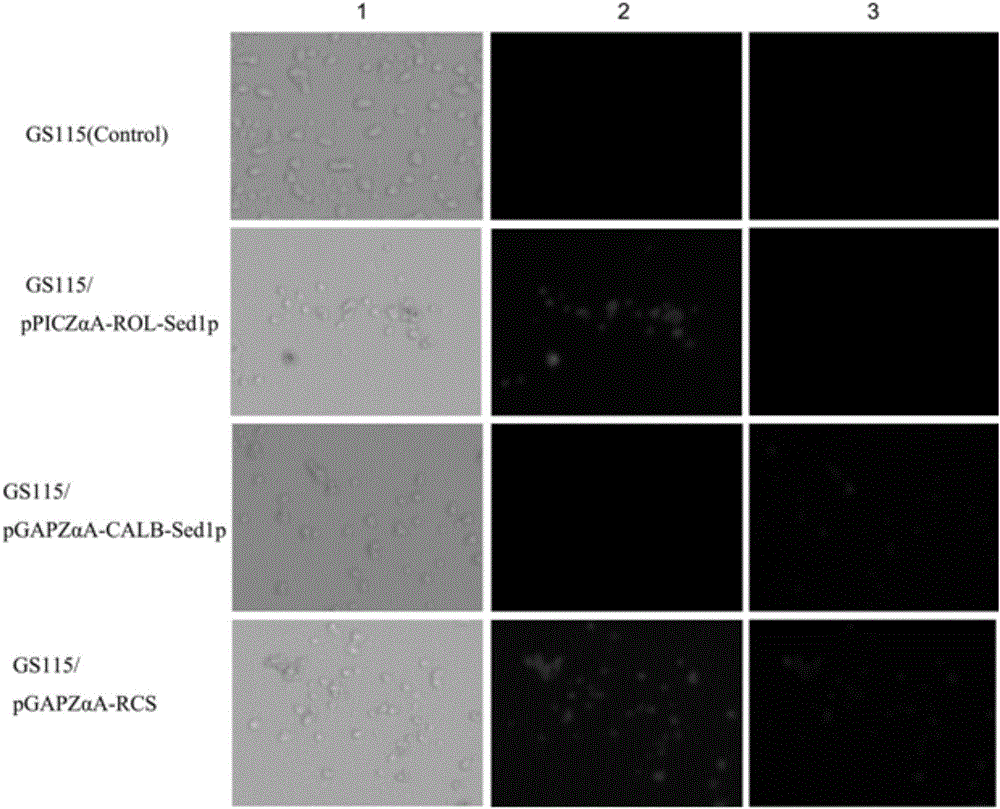
[0044] The construction of Rhizopus oryzae lipase (ROL) Pichia pastoris cell surface display system, the steps are as follows:
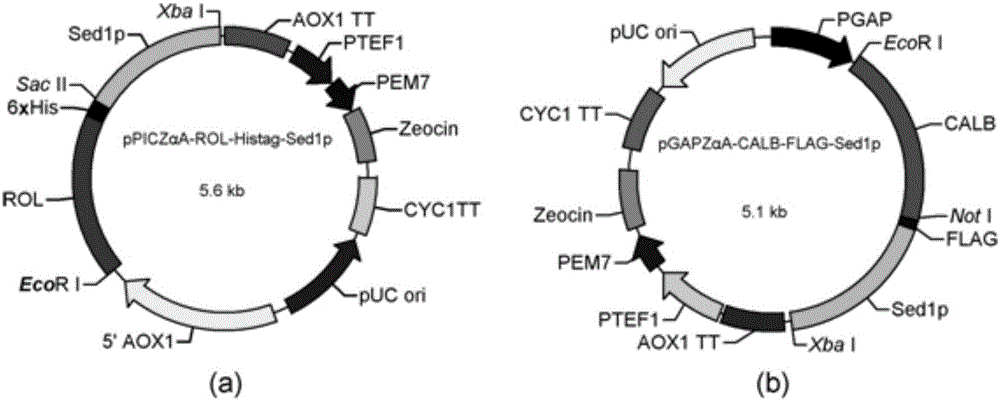
[0045] A. Construction of surface display recombinant plasmid vector pPICZαA-ROL
[0046] The lipase gene ROL from Rhizopus oryzae was cloned and fused with plasmid pPICZαA. The primers were designed according to the plasmid pPIC9K-ROL containing the lipase gene sequence of Rhizopus oryzae, and the primers were synthesized by Shanghai Sangong Bioengineering Co., Ltd.
[0047] R1 (5'-3', the same below): CCGGAATTCATGGTTCCTGTTTCTGGTAAATCTG, add EcoRI restriction site in front.
[0048] R2: TCCCCGCGG ATGATGATGATGATGATG CAAACAGCTTCCTTCGTTGATATCA, add SacⅡ restriction site and His-tag tag protein (underlined part) in front.
[0049] PCR amplification with synthetic primers, PCR reaction system: 1 μL plasmid pPIC9K-ROL, 1 μL R1 primer, 1 μL R2 primer, 25 μL PremixExtaq, 22 μL ddH 2 O. Amplification conditions were: denaturation at 94°C for 2min; dena...
Embodiment 3
[0061] The construction of Candida antarctica lipase B (CALB) Pichia pastoris surface display system, the steps are as follows:
[0062] A. Construction of surface display recombinant plasmid vector pGAPZαA-CALB
[0063] Cloning of the Candida antarctica lipase B gene CALB, fused to pGAPZαA. The primers were designed according to the lipase B gene sequence of Candida antarctica, and the primers were synthesized by Shanghai Sangon Bioengineering Co., Ltd.
[0064] C1: CCGGAATTCATGAAGCTACTCTCTCTGACCGGTG, with an EcoRI restriction site added in front.
[0065] C2: AAGGAAAAAAGCGGCCGCTGGGGGTGACGATGCCGGAGC, with a NotⅠ restriction site added in front.
[0066] Using Candida antarctica genomic DNA as a template, PCR amplification was performed with synthetic primers). PCR reaction system: 1 μL Candida antarctica genomic DNA, 1 μLC1 primer, 1 μLC2 primer, 25 μL PremixExtaq, 22 μL ddH 2 O. Amplification conditions were: denaturation at 94°C for 2min; denaturation at 94°C for 30s; ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com