High-flux nucleic acid analysis method and application thereof
A nucleic acid analysis, high-throughput technology, applied in the field of biotechnology and molecular diagnosis, can solve the problems of lack of detection methods, quantitative analysis, such as expression and copy number analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0150] Detect the typing of 48 SNP sites
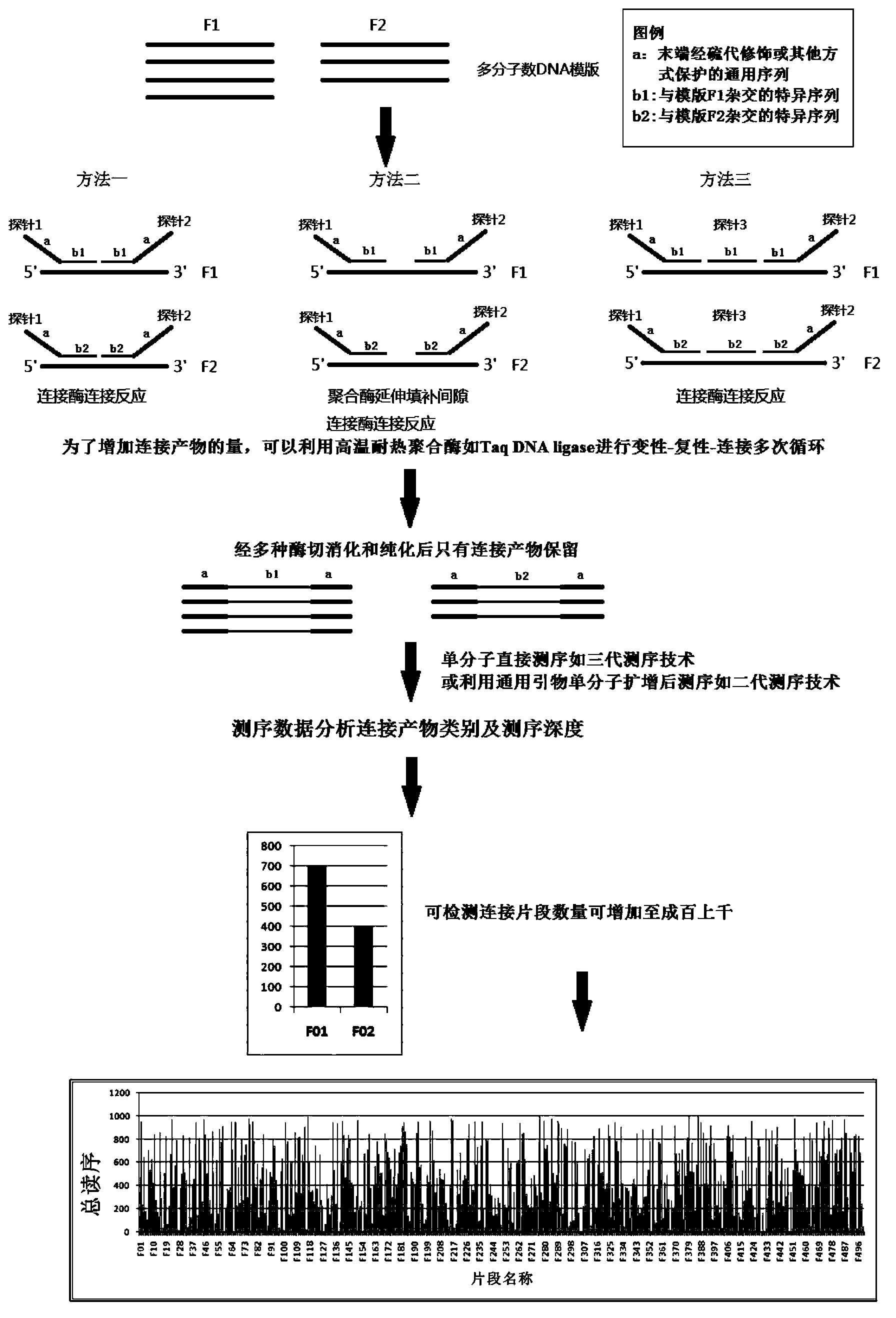
[0151] Design junction probes for 48 SNP sites, design 3 probes for each site, 2 5'-end allele-specific probes and 1 3'-end consensus sequence, the first half of the 5'-end probe A general PCR sequence compatible with the next-generation sequencing platform of Illumina is added in part, and another general PCR sequence compatible with the next-generation sequencing platform of Illumina is added to the second half of the 5' end probe. The probe was ligated under the action of TaqDNA ligase under the condition of good pairing with the template, and the ligation product was amplified using general PCR primers compatible with the illumina next-generation sequencing platform. Different samples were amplified with general primers with different tag sequences, and then After uniform mixing and purification, 1x72 sequencing was performed on the Illumina GAIIx sequencer. After the Sequencing reads are read out by software, different sample so...
Embodiment 2
[0180] Detection of DMD gene exon deletion duplication
[0181] Basic principles such as Figure 4 As shown, 141 probes were designed for each sample, 129 of which were distributed on the 79 exons of the DMD gene, 6 reference gene probes, and 6 sex chromosome sex identification probes (3 on the X chromosome, 3 on the Y chromosome). Two probes are designed for each site, one 5' end probe and one 3' end probe. For the specific sequence that hybridizes with the target nucleic acid fragment, the 5' end of the 3' end probe is modified by phosphorylation, the first half is the specific sequence that hybridizes with the target nucleic acid fragment, and the second half is the general sequence consistent with the subsequent PCR amplification primers . The probes were ligated under the action of Taq DNA ligase under the condition of good pairing with the template, and the ligation products were amplified with universal PCR primers compatible with the Illumina next-generation sequenc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com