Detection method for biomolecular probe-calibrated specific sites of DNA based on nanopore device
A technology of biomolecular probes and specific sites, applied in biochemical equipment and methods, measurement/inspection of microorganisms, electrochemical variables of materials, etc., can solve the problem of high computational complexity and algorithm complexity, reduce time efficiency, and increase sequencing Cost and other issues, to achieve the effect of improving time resolution, reducing workload, and maintaining the signal-to-noise ratio of current signals
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0039] 1) Preparation of chip devices
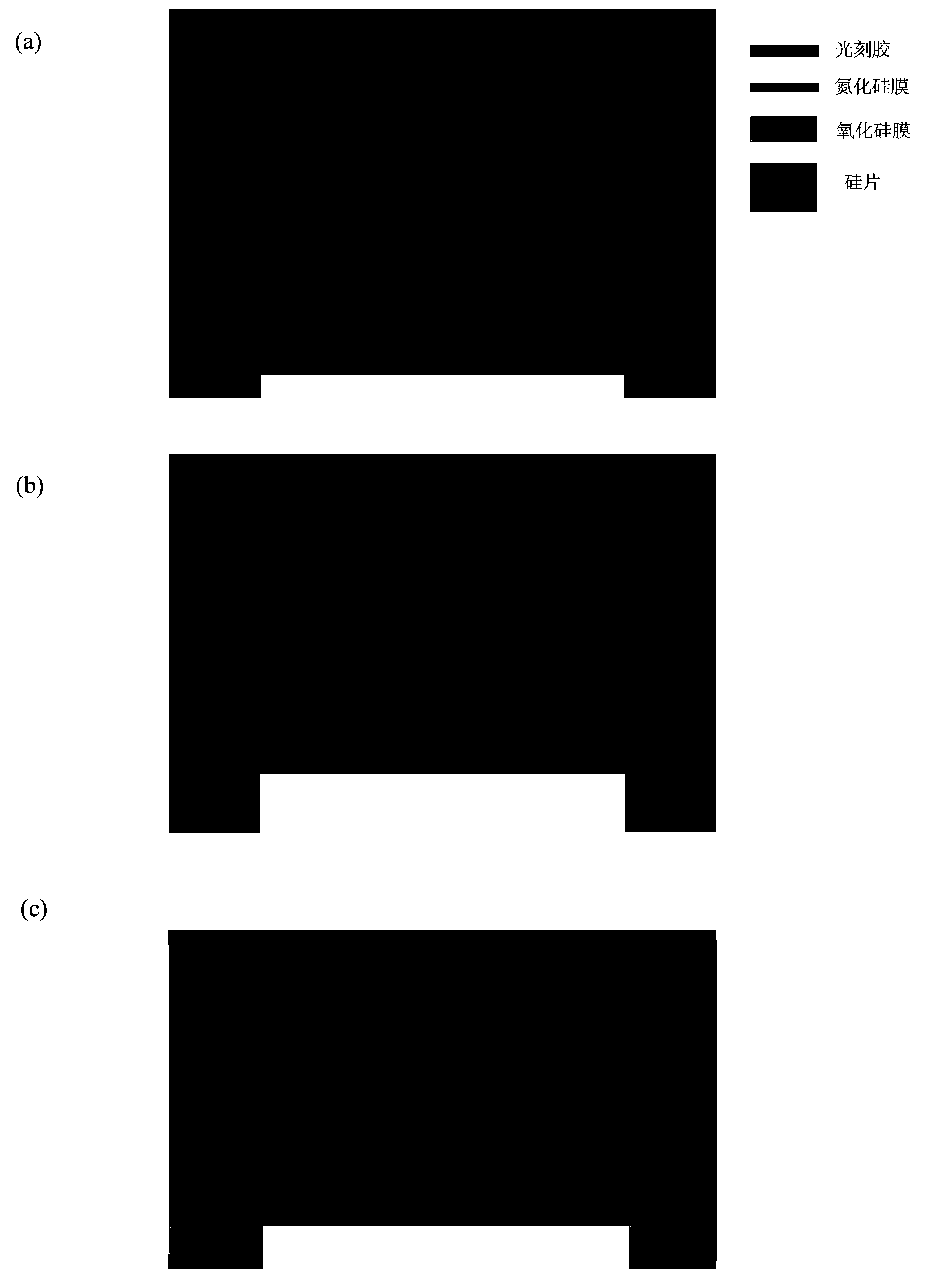
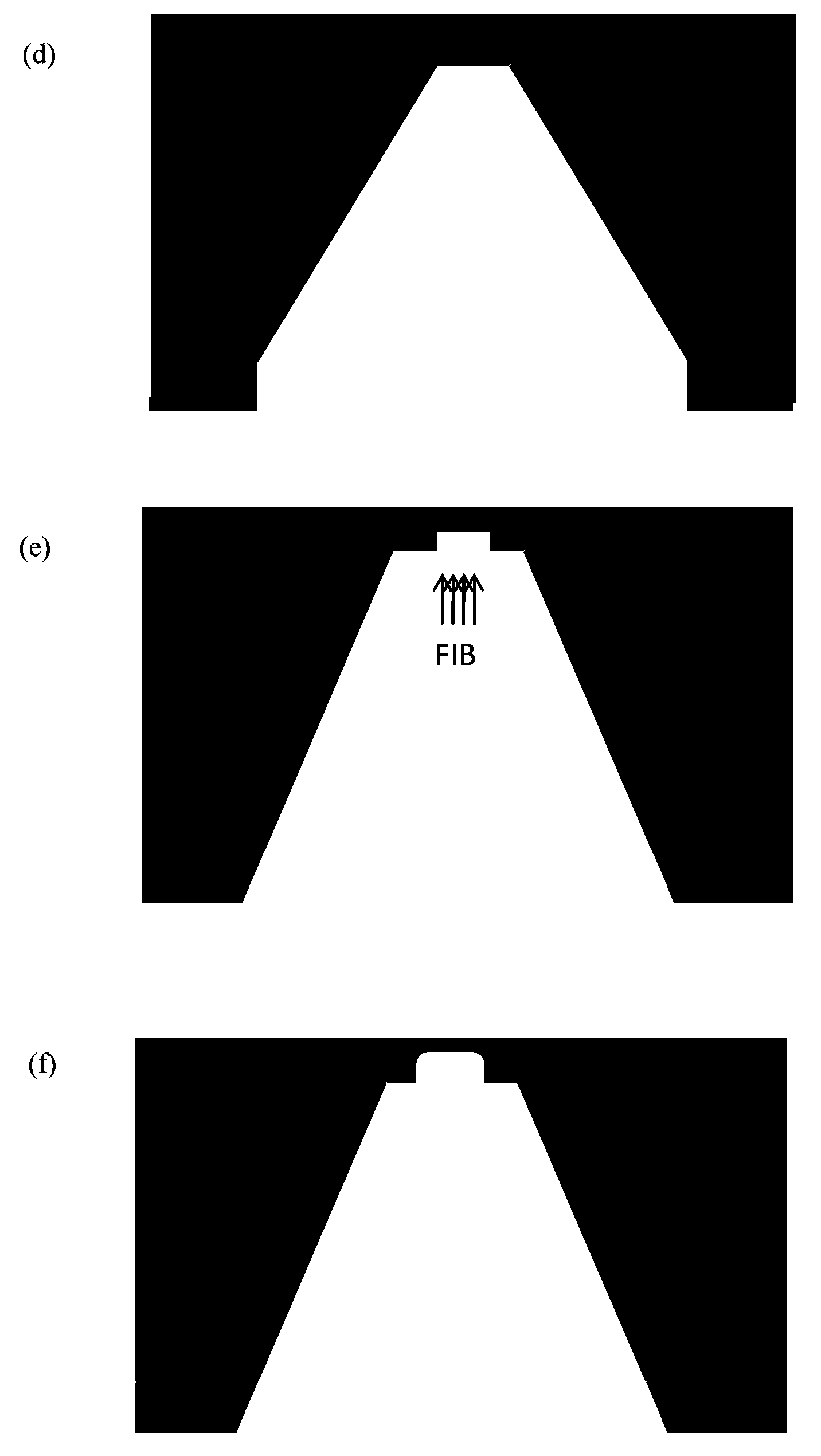
[0040] The purpose of this step is to prepare a solid-state nanoporous device that can be operated in a transmission electron microscope, so that the high-energy convergent electron beam in the transmission electron microscope can be used to punch holes in the suspended film of the device, thereby realizing a nanoporous device. This includes a series of micro-nano processing technology involved in traditional semiconductor processing technology, and also involves a series of modern cutting-edge nano-scale processing technology.
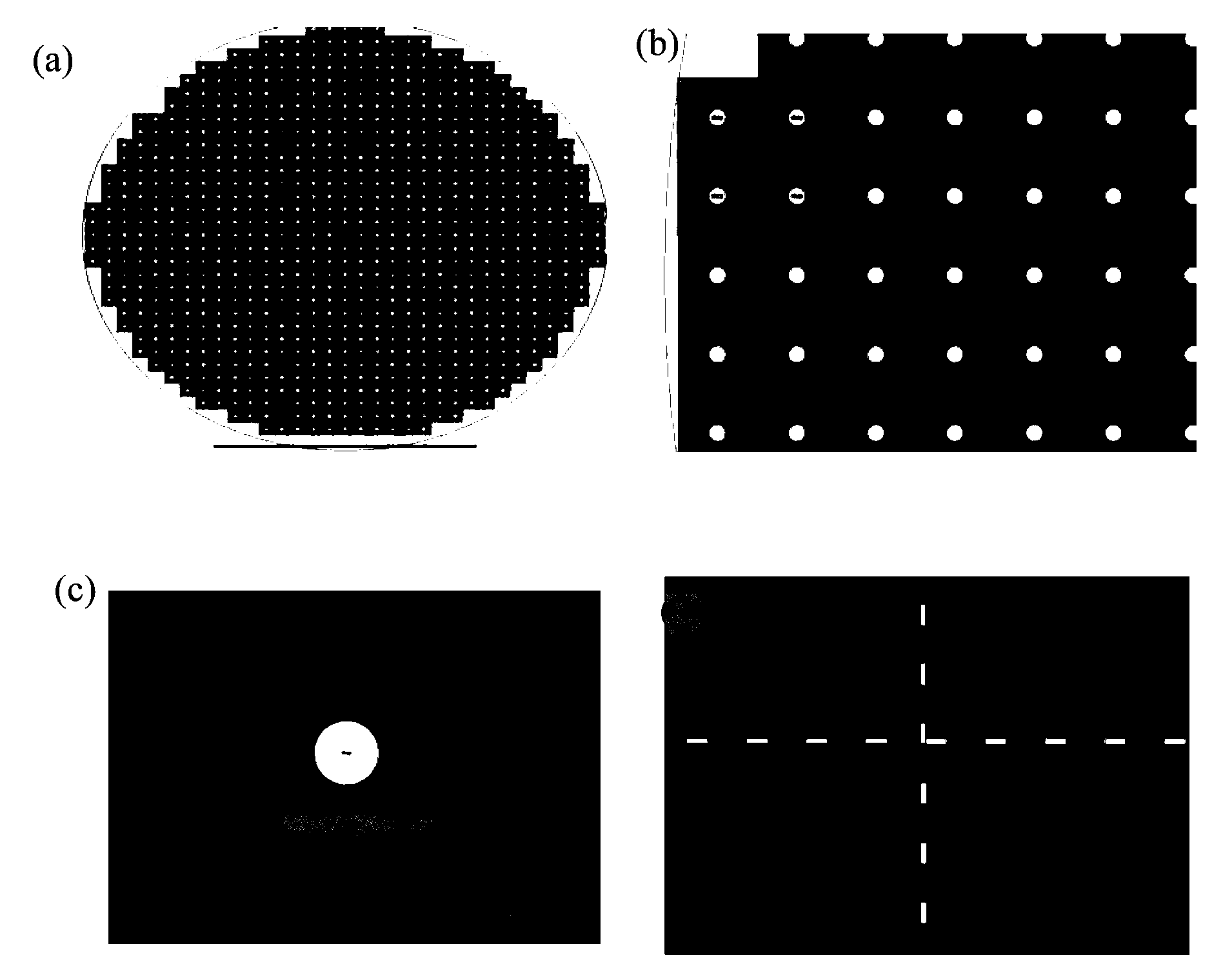
[0041] The specific method is as follows: First, a 4-inch silicon wafer with a (100) surface is grown with 2-micron silicon oxide on both sides, and low-stress silicon nitride of about 150 to 200 nanometers is deposited by low-pressure chemical vapor deposition. Then make a photolithography mask, the purpose is to make a lot of 3mm×3mm small substrate periodic distribution on a 4-inch silicon wafer ( figure 1 a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| depth | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com