Single-cell genome analysis method and kit
A genome analysis and single-cell technology, which is applied in the field of biological single-cell genome research, can solve the problem of less sequencing of single-cell genomes, and achieve the effect of simple operation and avoiding waste
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
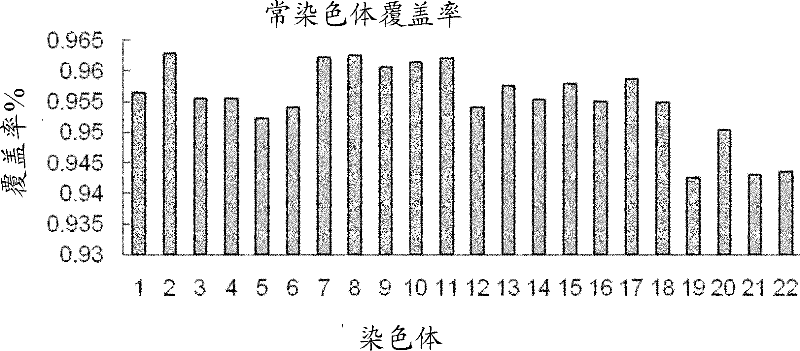
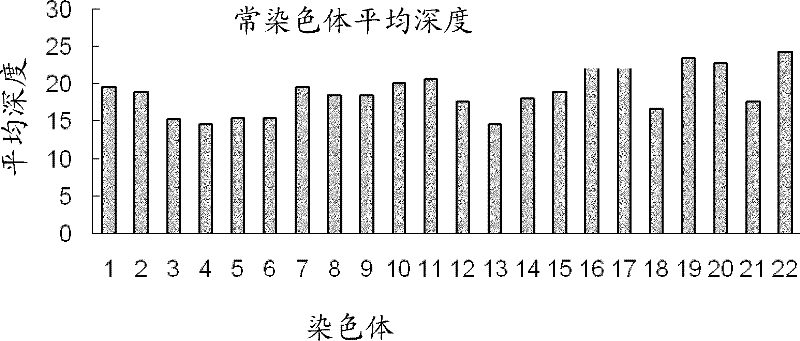
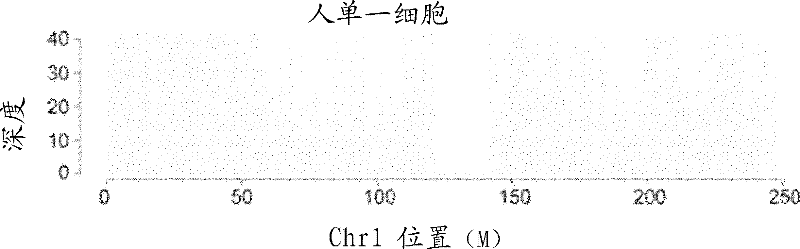
[0071] Single-cell genome studies in human cell lines
[0072] (1) Separation and lysis of single cells: add the immortalized human lymphocyte line single cells to the PBS droplet on the culture dish, dilute appropriately (that is, dilute according to the concentration of the cell line cells, until observed in the 200X field of view of the microscope until the number of cells is between 10-20), single cells were separated by oral suction under a microscope, and the obtained single cells were placed in a PCR tube containing 1.5-2uL ALB (Alkaline Lysis Buffer, specific formula: 50mMDTT, 200mM KOH), Place at -20°C to -80°C for at least 30min.
[0073] Heat the PCR tube containing single cells at 62°C-68°C, preferably 65°C, for 8-12min to lyse the cells and release the chromosomal DNA.
[0074] (2) Whole Genome Amplification (WGA):
[0075] Any of the following methods of multiple displacement amplification (MDA) or DOP-PCR whole-genome amplification can be used.
[0076] ①Multip...
Embodiment 2
[0103] Human Housekeeping gene primers for PCR detection of single cell WGA products
[0104] The single cells isolated from human tissue or blood are lysed and treated with WGA (refer to Example 1 for the specific treatment process), and the products are detected by Housekeeping Gene PCR.
[0105] The selected Housekeeping Gene and its corresponding primer information are as follows:
[0106]
[0107]
[0108] The aforementioned Housekeeping Gene is described in Eli Eisenberg and Erez Y. Levanon, (2003) Human housekeeping genes are compact. Trends in Genetics. 19(7): 362-365. The inventors found that, compared with other Housekeeping Genes on other chromosomes, the above selected Housekeeping Gene and its primers can be conveniently and accurately used for the qualitative detection of WGA amplification products in human cells.
[0109] The PCR system contains: heat-resistant DNA polymerase with 3'exolytic activity; single-cell WGA product (template); dNTP mix; Mg 2+ ;...
Embodiment 3
[0132] Genome Study of Pollen Cells of P. amabilis
[0133] This technique was applied to the genome research of P. amabilis pollen cells. It is intended that after the genome is amplified, the sample volume is sufficient to construct the Solexa DNA library, and finally sequenced on the machine.
[0134] Considering that there are 3 to 5 single cells in the pollen grains of plants, first separate the pollen grains one by one under an inverted microscope, add a mixed solution of 4% cellulase and 2% pectinase equal to the volume of the pollen grains, and place Mix for 6 hours at room temperature in the dark on a vertical mixer to lyse and break the cell wall. After completion, add the lysis system of the same pollen grain to the PBS droplet on the petri dish, separate single cells under the microscope, and put the obtained single cells into a PCR tube containing 1.5uL ALB (alkaline lysis buffer).
[0135] Subsequently, the MDA (multiple displacement amplification) reaction was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com