High temperature alkaline xylanase XYN10A, gene thereof and application thereof
A technology of xylanase and mannanase, which is applied in the field of genetic engineering, can solve the problems of inability to adapt to alkaline environment, loss, low expression amount and limit the prospect of industrial production, and achieves excellent pH stability, high activity, enzyme live stable effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0102] Cloning of Example 1 Xylanase Encoding Gene Xyn10A
[0103] Extraction of Humicola sp.S8 genomic DNA:
[0104] After 3 days of culture, the mycelium was centrifuged at high speed and taken into a mortar, frozen in liquid nitrogen and ground for 5 minutes, then the grinding liquid was placed in a 50mL centrifuge tube, 2mL of CTAB extract was added, and lysed in a water bath at 70°C for 2h, every 10min Mix once and centrifuge at 12000rpm for 10min at 4°C. The supernatant was extracted in phenol / chloroform to remove impurity proteins, and then an equal volume of isopropanol was added to the supernatant. After standing at room temperature for 10 minutes, centrifuge at 12,000 rpm for 10 minutes at 4°C. The supernatant was discarded, the precipitate was washed twice with 70% ethanol, dried in vacuo, dissolved in 0.2 mL TE, and stored at -20°C for later use.
[0105] The degenerate primers P1 and P2 were designed and synthesized according to the conserved (WDVVNE and NDY(F)N...
Embodiment 2
[0113] The preparation of embodiment 2 recombinant xylanase
[0114] The expression vector pPIC9 is subjected to double enzyme digestion (SpeI+NotI), and at the same time, the amplified xyn10A gene encoding xylanase (without signal peptide) is subjected to double enzyme digestion (EcoR I+Not I), and the xylan encoding xylan is excised The gene fragment of the enzyme was connected with the expression vector pPIC9 to obtain the recombinant plasmid pPIC9-xyn10A containing the xylanase gene xyn10A and transform Pichia GS115 to obtain the recombinant Pichia strain GS115 / xyn10A; The cDNA of xylanase XYN10A was inserted into the expression vector pPIC9 from which the α-factor signal peptide sequence had been removed by enzyme digestion and ligation, and the recombinant plasmid pPIC-xyn10A-1 containing the gene xyn10A encoding xylanase sequence containing the signal peptide sequence was obtained And transform Pichia pastoris GS115 to obtain recombinant Pichia pastoris strain GS115 / xyn...
Embodiment 3
[0116] The activity analysis of embodiment 3 recombinant xylanase
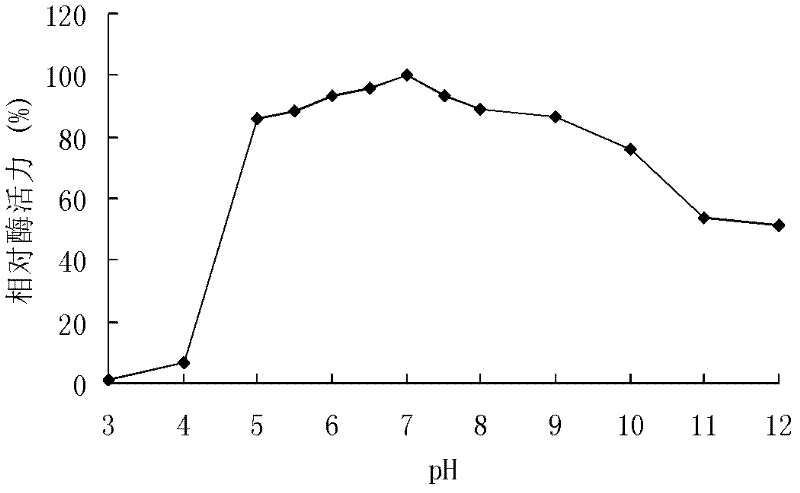
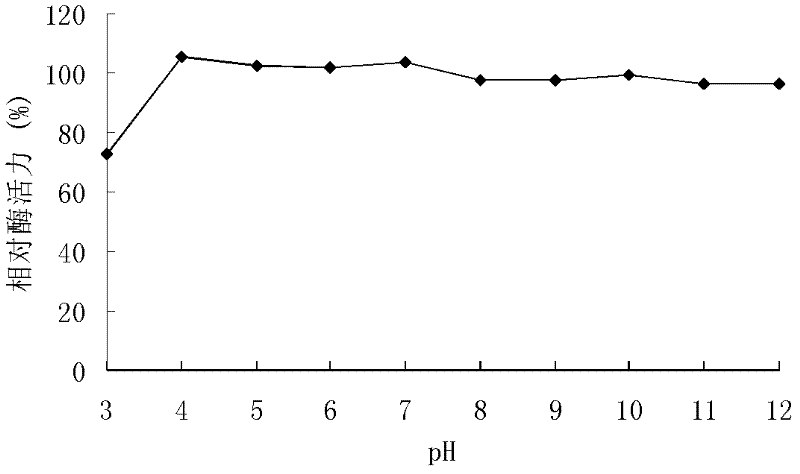
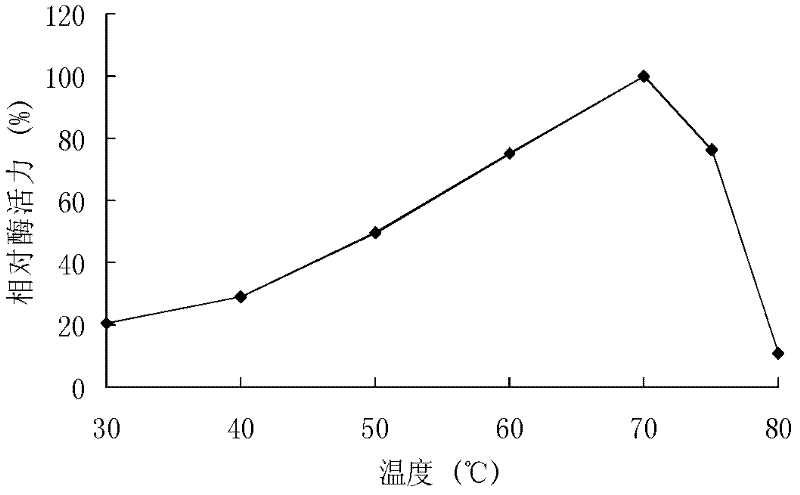
[0117] The recombinant xylanase produced by recombinant strains GS115 / XYN10A and GS115 / XYN10A-1 was purified, and the activity was analyzed by DNS method.
[0118] DNS method: The specific method is as follows: at pH 7.0, 60°C, 1 mL of reaction system includes 100 μL of appropriate diluted enzyme solution, 900 μL of substrate, reacted for 10 minutes, added 1.5 mL of DNS to terminate the reaction, and boiled for 5 minutes. After cooling, the OD value was measured at 540 nm. One enzyme activity unit (U) is defined as the amount of enzyme that releases 1 μmol of reducing sugar per minute under given conditions.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com