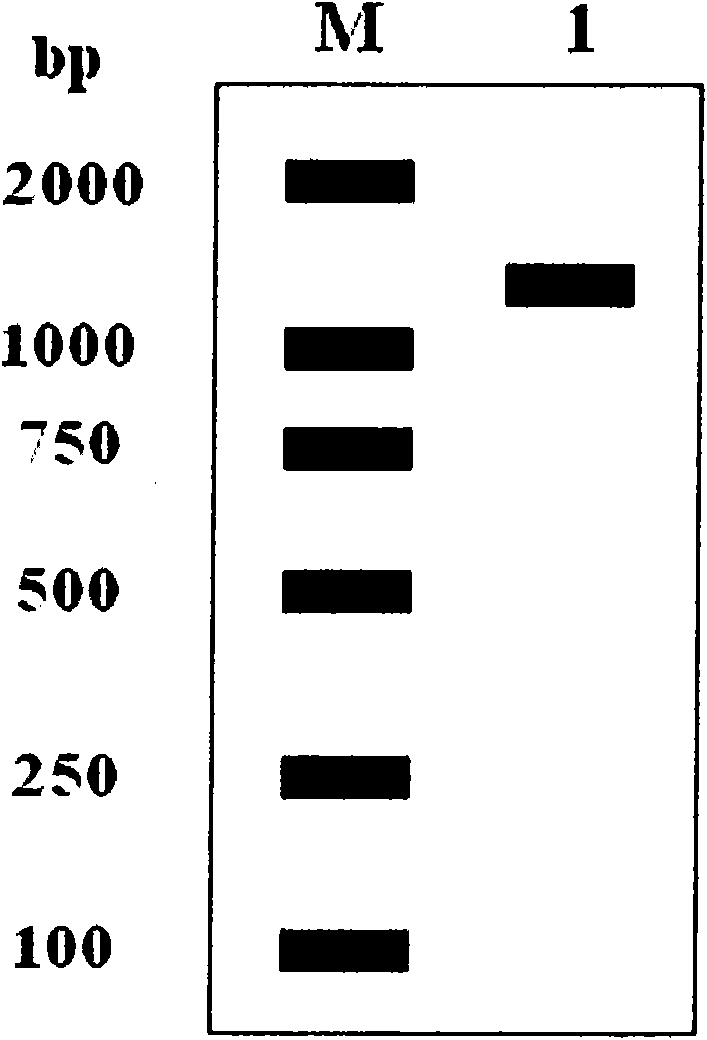Tea tree cytoplasm-type glutamine synthetase (GS) gene and encoding protein thereof
A technology of glutamine and synthetase, applied in the field of genetic engineering, can solve the problems such as there is no GS gene clone report of tea tree yet
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Extraction of tea total RNA and acquisition of cDNA.
[0035] The total RNA of tea tree (Longjing 43) was extracted by plant total RNA purification kit (MACHEREY NAGEL company), and the total cDNA was obtained by using the extracted tea tree total RNA by Reverse Transcription System kit (Promega company), and the total cDNA was stored at -20°C. save.
Embodiment 2
[0036] Example 2: Obtaining the complete sequence of glutamine synthetase.
[0037] Design upstream and downstream primers cyGS-5' and cyGS-3':
[0038] cyGS-5' sequence: GAAAACAAACACACAGATAATATAA,
[0039] cyGS-3' sequence: TCATGGTTTCCACAGGATGGTGGTAG.
[0040] The total cDNA of tea tree was used as the template for PCR amplification. The PCR reaction program was: pre-denaturation at 95°C for 3 minutes; denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 1 minute and 30 seconds, and 35 cycles of reaction; extension at 72°C for 10 minutes; storage at 4°C . 1% agarose gel electrophoresis to separate the PCR products, AxyPrep DNA Gel Recovery Kit (AXYGEN Company) to recover the PCR products, connected to the pMD19-T vector (TaKaRa Company), connected at 16°C overnight, and transferred the connection solution to the large intestine In E. coli DH5α competent cells, pour LB solid medium (containing Amp) and evenly spread 40 μl of 5-bromo-4...
Embodiment 3
[0043] Embodiment 3: Construction of pET28b-GS recombinant plasmid
[0044] Design primers:
[0045] cyGS-F1: CCATGGGCATGCATCATACTGAATCATCGTCGT;
[0046] cyGS-F2: CTCGAGAACACCCAACTGGTTTTGCACC, Nco I and Xho are underlined respectively
[0047] I restriction site.
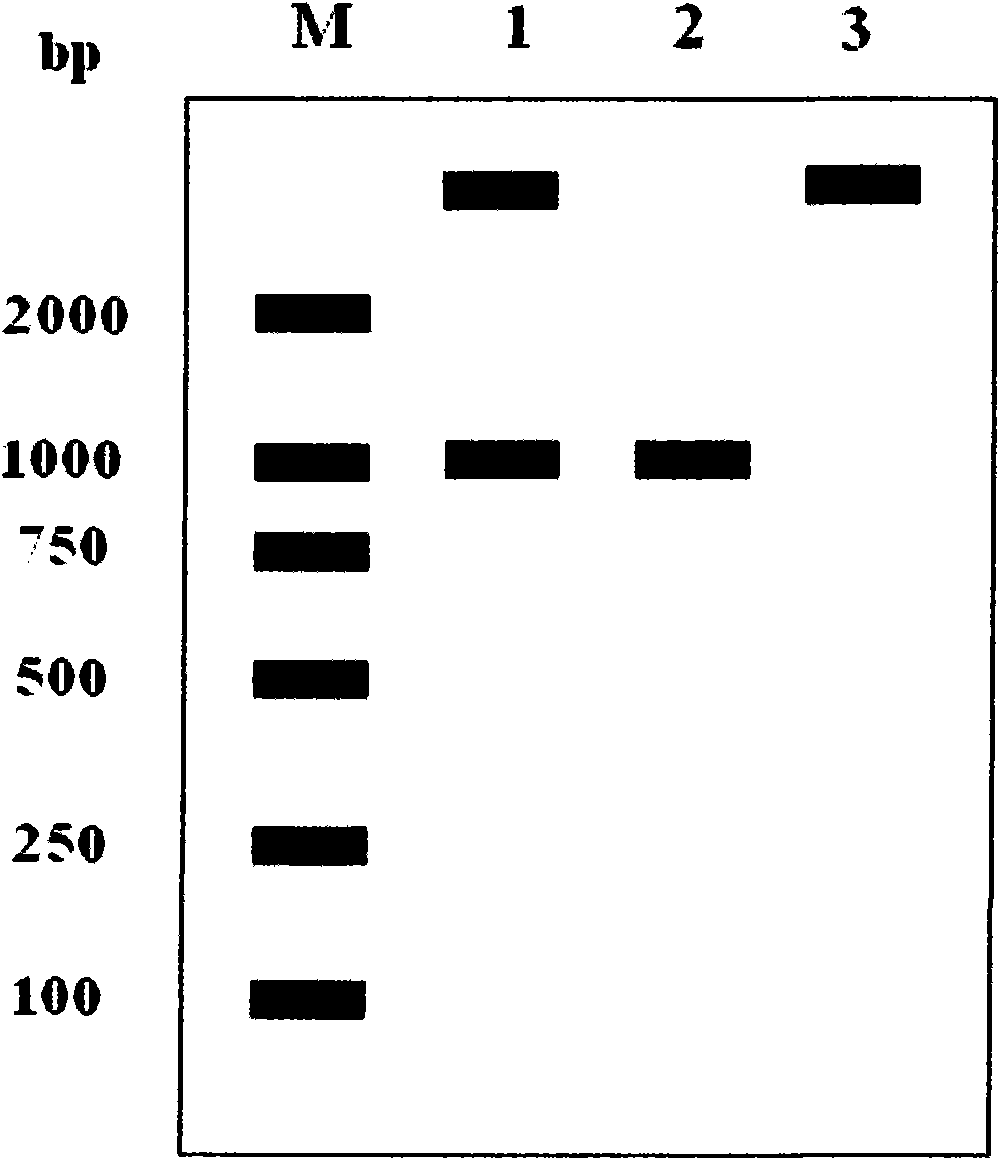
[0048] Using the full sequence of the Cam-cyGS gene in Example 1 as a template, the protein coding region was amplified by PCR. The reaction conditions were: pre-denaturation at 95°C for 3 minutes; denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 1 minute and 10 seconds, and 35 cycles of reaction ; Extend at 72°C for 10 minutes; Store at 4°C. Amplified product ( figure 2 ) and vector pET28b were digested with Nco I and Xhol I for 4 hours, connected and transformed into E. coli DH5α competent cells, poured on LB plates, screened positive clones, and obtained expression plasmid pET28b-GS. After identification by Nco I and XhoI double enzyme digestion ( image 3 ), sen...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Relative molecular mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com