DNA sequence, recombinant vector, single and double auxotrophic Hansenula polymorpha, and preparation method thereof
A technology of Hansenula polymorpha and auxotrophy, applied in the field of genetic engineering, can solve the problems of high drug resistance and false positive ratio, difficulty in obtaining transformants, unclear mutation sites, etc., and achieve convenient source of strains. , The effect of clear mutation mechanism and low reversion mutation rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
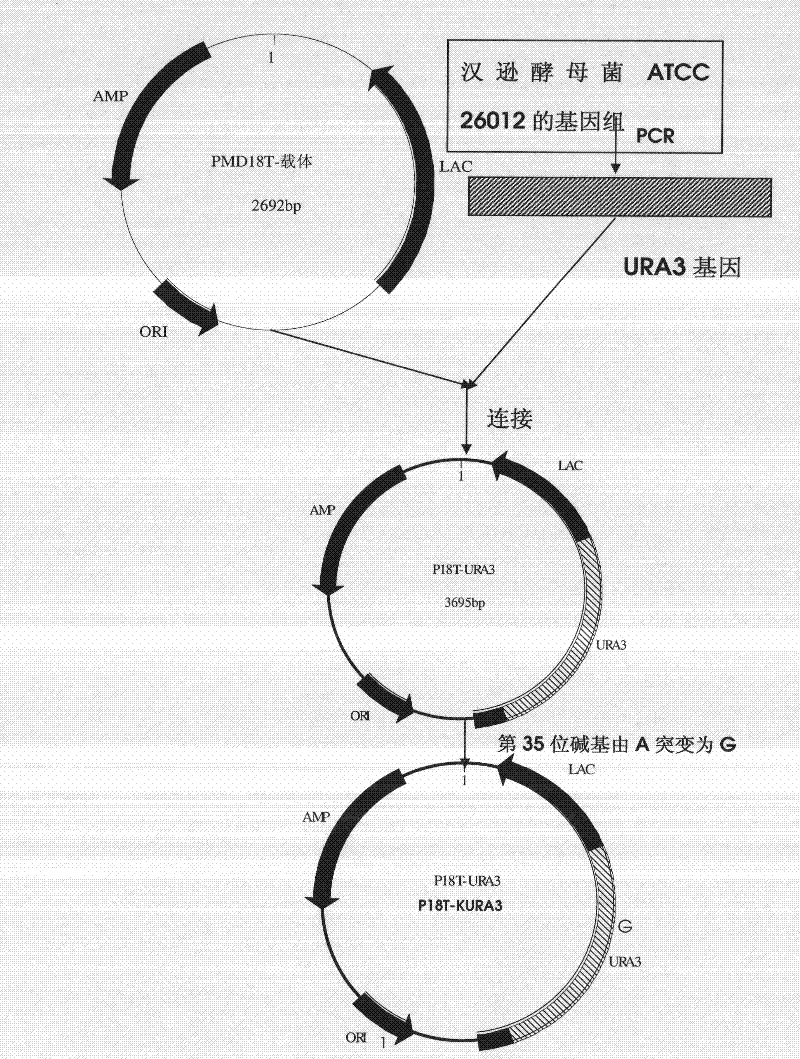
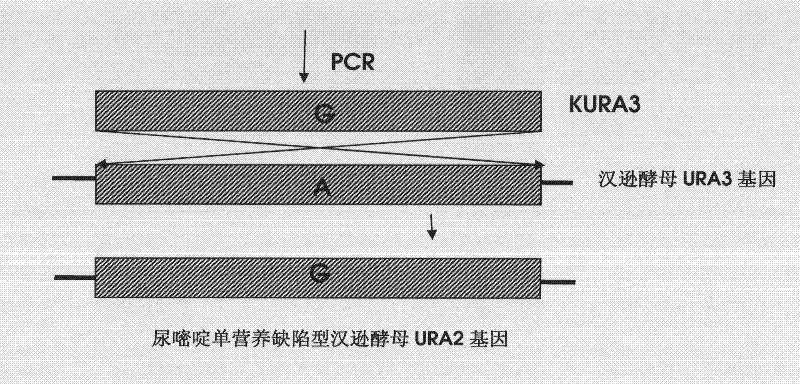
[0097] The construction of the recombinant plasmid p18T-KURA3 of embodiment 1 inactivation URA3 gene
[0098] Such as figure 1 As shown, the construction process of the recombinant plasmid p18T-KURA3 is as follows. The primers were designed according to the reported (GenBank) nucleotide sequence of URA3 of Hansenula polymorpha, as shown in Table 1.
[0099] Table 1
[0100]
[0101] Extract the total DNA of Hansenula polymorpha ATCC 26012 as a template, use URA3-F and URA3-B as primers, and amplify the URA3 gene sequence according to the following PCR reaction system:
[0102] Template DNA 2μl
[0103] URA3-F 1μl
[0104] URA3-B 1μl
[0105] dNTP 5μl
[0106] 10*PCR ExTaq buffer 5μl
[0107] ExTaq enzyme 1μl
[0108] Deionized water 35μl
[0109]
[0110] Total volume 50μl
[0111] Wherein, the reaction condition of carrying out PCR reaction on PCR instrument is as follows:
[0112] 94°C, 5 minutes; then cycle 30 times at 94°C, 40 se...
Embodiment 2
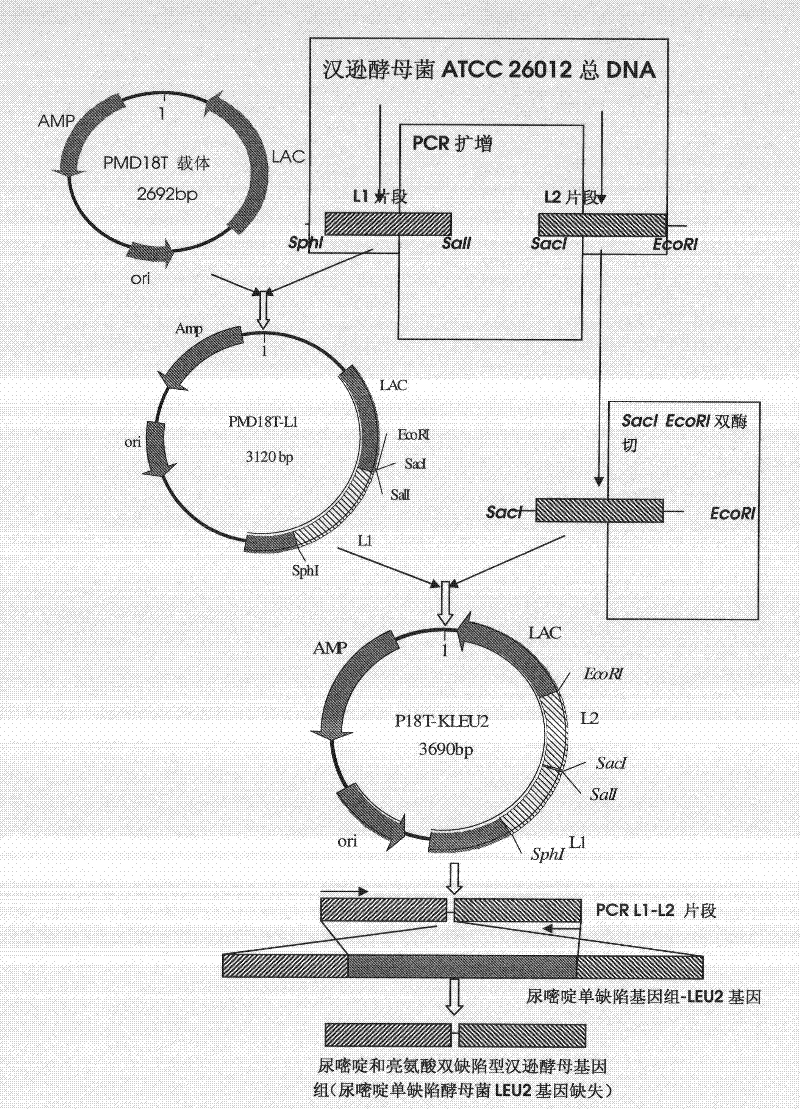
[0134] Example 2 Construction of the recombinant plasmid p18T-KLEU2 containing the inactivated LEU2 gene
[0135] Such as figure 2 As shown, the construction process of the recombinant plasmid P18T-KLEU2 is as follows.
[0136] Primers were designed according to the reported (GenBank) nucleotide sequence of Hansenula polymorpha LEU2, see Table 2.
[0137] Table 2
[0138]
[0139] The total DNA of Hansenula polymorpha ATCC 26012 was extracted as a template, and a segment of upstream DNA sequence of LEU2 gene, namely L1 fragment, was amplified with LEU2-F1 and LEU2-B1 as primers, with a length of 525 bp. LEU2-F2 and LEU2-B2 were used as primers to amplify a downstream DNA sequence of the LEU2 gene, namely the L2 fragment, with a length of 570 bp.
[0140] Proceed according to the following PCR reaction system, wherein primer 3 is LEU2-F1, primer 4 is LEU2-B1 or primer 3 is LEU2-F2, primer 4 is LEU2-B2:
[0141] Template DNA 2μl
[0142] Primer 31μl
[0143] Primer 41μ...
Embodiment 3
[0153] Example 3 Construction and identification of uracil monoauxotrophic Hansenula polymorpha
[0154] The p18T-KURA3 constructed in Example 1 was transformed into Hansenula polymorpha ATCC 26012 by electroporation, and the uracil monoauxotrophic Hansenula strain was screened and identified. The specific steps were as follows:
[0155] 1. Preparation of competent Hansenula polymorpha ATCC 26012
[0156] Hansenula polymorpha ATCC 26012 was streak-inoculated on the slant medium (it is the YPD medium containing 7.5% agar) for activation, and the monoclonal colony was picked and inoculated in 2ml of YPD liquid medium with an inoculation loop , at 37°C in a shaker at 250rpm for 24 hours, then inoculate 2ml of the bacterial liquid into 50ml of YPD liquid medium, and cultivate it in a shaker at 250rpm at 37°C until the logarithmic growth phase (OD 600 Between 1.2-1.5), centrifuge at 3000rpm for 10 minutes and collect the bacteria; resuspend with potassium phosphate buffer of pH 7....
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com