Chinese hamster microsatellite genetic marker and screening method thereof
A technology of microsatellite marker and genetic marker, applied in the field of Chinese hamster microsatellite genetic marker and its screening
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
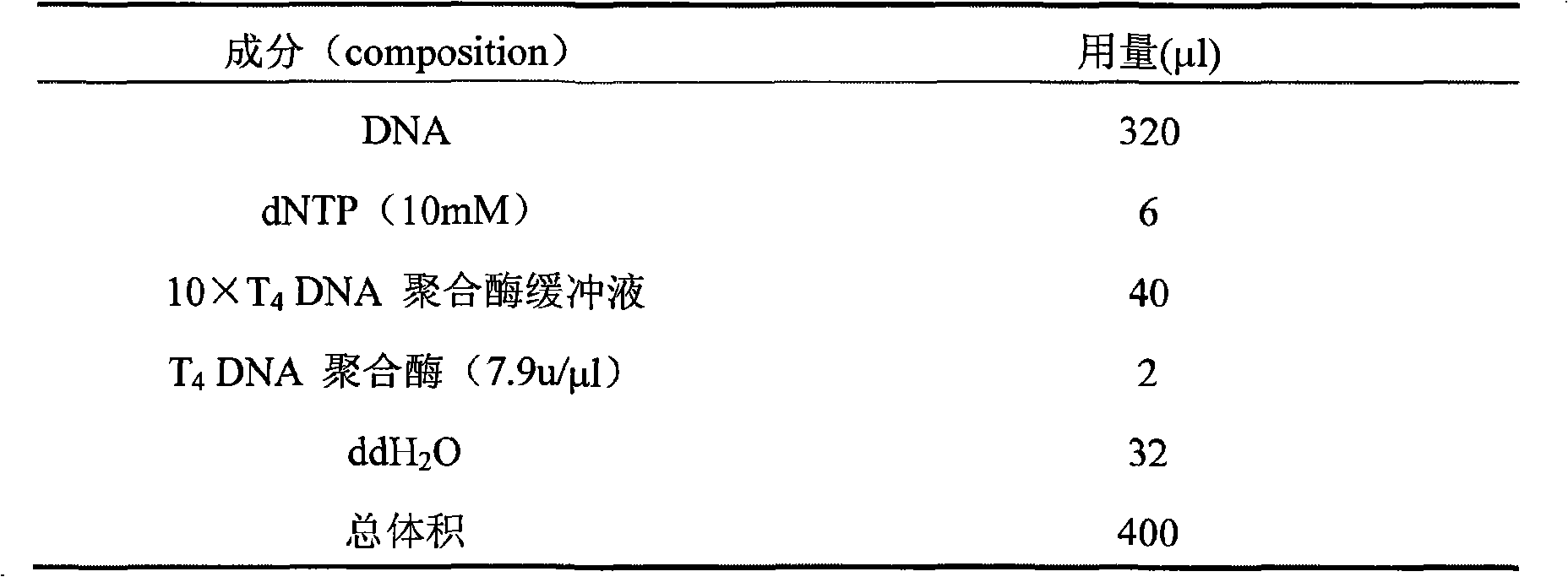
[0011] 1. Construction of Chinese hamster genome microsatellite enrichment library
[0012] 1. DNA extraction
[0013] (1) Take Chinese hamster tail tissue, cut it into pieces with scissors, add 500 μl of lysis buffer, mix well, add proteinase K to a final concentration of 100L, mix well, and digest overnight at 50°C on a slow shaker.
[0014] (2) Cool the solution to room temperature, add an equal volume of Tris saturated phenol, and slowly invert until the two phases are mixed to form an emulsion. Centrifuge at 13200 / rpm at 4°C for 10 minutes, and slowly transfer the supernatant to another centrifuge tube with a 1ml pipette tip.
[0015] (3) Add an equal volume of Tris saturated phenol and chloroform, shake well, centrifuge at 13200 / rpm for 10 min at 4°C, and transfer the supernatant to another centrifuge tube.
[0016] (4) Add an equal volume of chloroform, shake well, centrifuge at 13200 / rpm for 10 min at 4°C, and transfer the supernatant to another centrifuge tube.
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com