Expression of enzymes involved in cellulose modification
a technology of enzymes and cellulose, applied in the field of expression of enzymes involved in cellulose modification, can solve the problem that the content of plastids in the tissue may not be sufficient for chloroplast transformation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
Construct Preparation
Constructs and methods for use in transforming the plastids of higher plants are described in Zoubenko et al. (Nuc Acid Res (1994) 22(19):3819-3824), Svab et al. (Proc. Natl. Acad. Sci. (1990) 87:8526-8530 and Proc. Natl. Acad. Sci. (1993) 90:913-917) and Staub et al. (EMBO J. (1993) 12:601-606). The complete DNA sequences of the plastid genome of tobacco are reported by Shinozaki et al. (EMBO J. (1986) 5:2043-2049). All plastid DNA references in the following description are to the nucleotide number from tobacco.
A vector was prepared to direct the expression of the Acidothermus E1 .beta.-1,4-endoglucanase in plant plastids. The plasmid pMPT4, a pGEM (Clonetech) derivative containing the entire Acidothermus E1 cellulase coding sequence (U.S. Pat. No. 5,536,655) and flanking regions on a 3.7 kb Pvu I genomic DNA fragment, was digested with the restriction endonuclease sites SacII and Asp718 to remove the coding sequence for the mature E1 cellulase protein. This f...
example 2
Tobacco plants transformed to express T7 polymerase from the nuclear genome and targeted to the plant plastid are obtained as described in McBride et al U.S. Pat. No. 5,576,198. Transgenic tobacco plants homozygous for the plastid targeted T7 polymerase are used for plastid transformation using particle bombardment.
Tobacco plastids are transformed by particle gun delivery of microprojectiles. Since integration into the plastid genome occurs by homologous recombination and the target site is between the acetyl CoA carboxylase and the large subunit of RuBisCo (rbcL), a single copy of the transgene is expected per plastid genome (Svab et al. (1993) supra).
Tobacco seeds (N. tabacum v. Xanthi N / C) homozygous for pCGN4026 (McBride et al., U.S. Pat. No. 5,576,198) T-DNA are surface sterilized in a 50% chlorox solution (2.5% sodium hypochlorite) for 20 minutes and rinsed 4 times in sterile H.sub.2 O. The seeds are then plated asceptically on a 0.2.times. MS salts...
example 3
Analysis of Cellulase Expression in Plastids
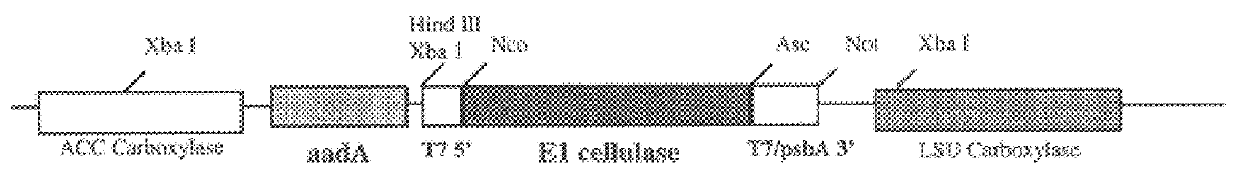
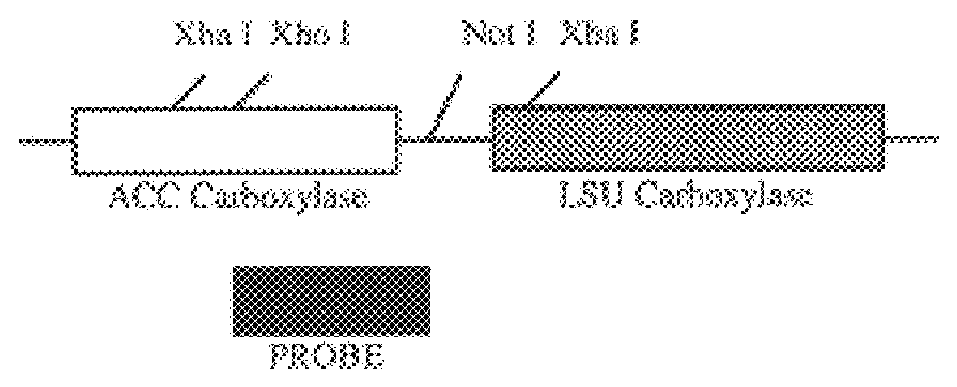
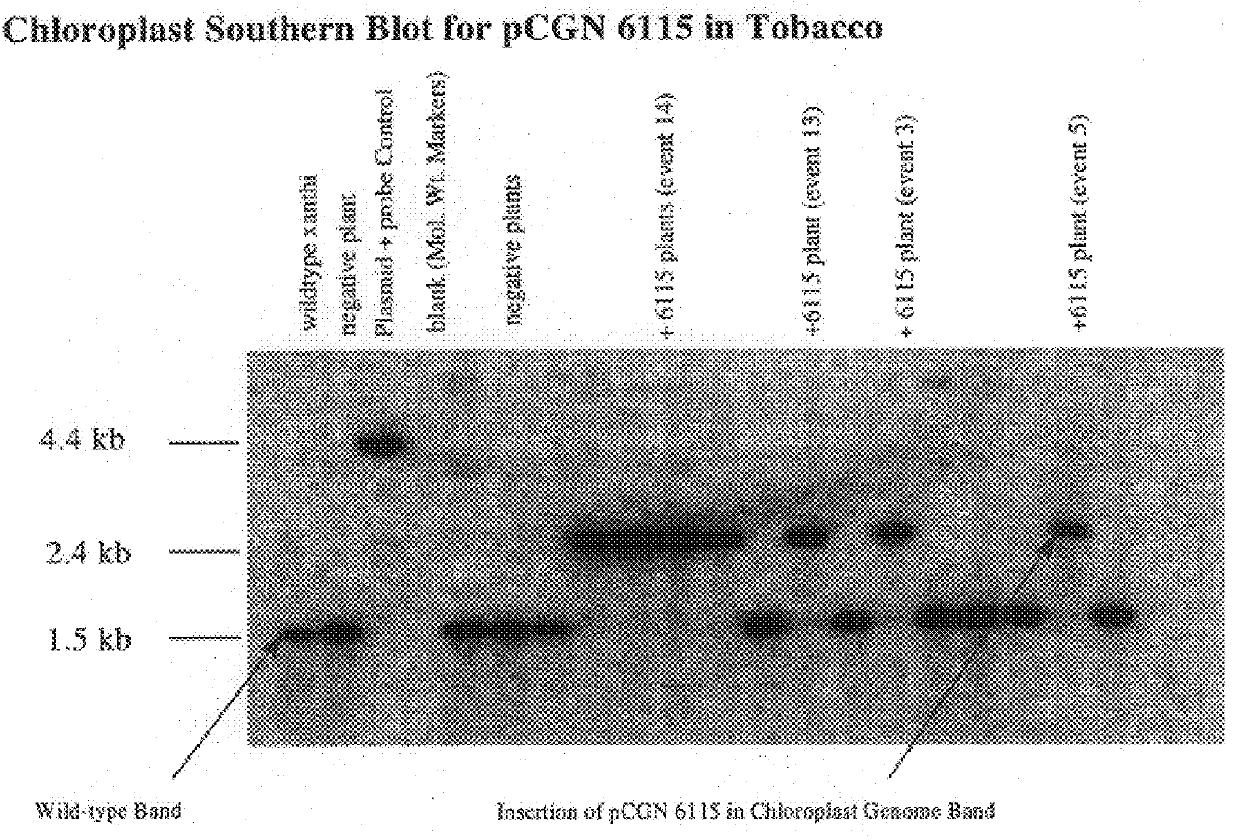
Following plastid transformation as described above, five independently isolated homoplasmic lines generated in the nuclear encoded T7 RNA polymerase producing background were generated. A schematic of pCGN6115 construct and a representation of incorporation into the tobacco plastid genome is shown in FIG. 1. The upper line represents the incoming DNA donated from pCGN6115 and the middle line represents the integration target region. Expected sizes for XbaI fragments are shown for the incoming DNA as well as for wild type DNA. As there is no XbaI site on the 5' end of the incoming DNA the combined size of the two chimeric genes is indicated. Also shown in FIG. 1 is the location of the probe used for Southern analysis. Homoplasmy was determined by Southern blot analysis as shown in FIG. 2.
To confirm homoplasmy by Southern hybridization, total plant cellular DNA is prepared as described by Bernatzky and Tanksley ((1986) Theor Appl Genet. 72:...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperatures | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com