Novel adenovirus
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
of ChAd155
[0417]Wild type chimpanzee adenovirus type 155 (ChAd155) was isolated from a healthy young chimpanzee housed at the New Iberia Research Center facility (New Iberia Research Center; The University of Louisiana at Lafayette) using standard procedures as described in Colloca et al. (2012) and WO2010086189, which is hereby incorporated by reference for the purpose of describing adenoviral isolation and characterization techniques
example 2
ector Construction
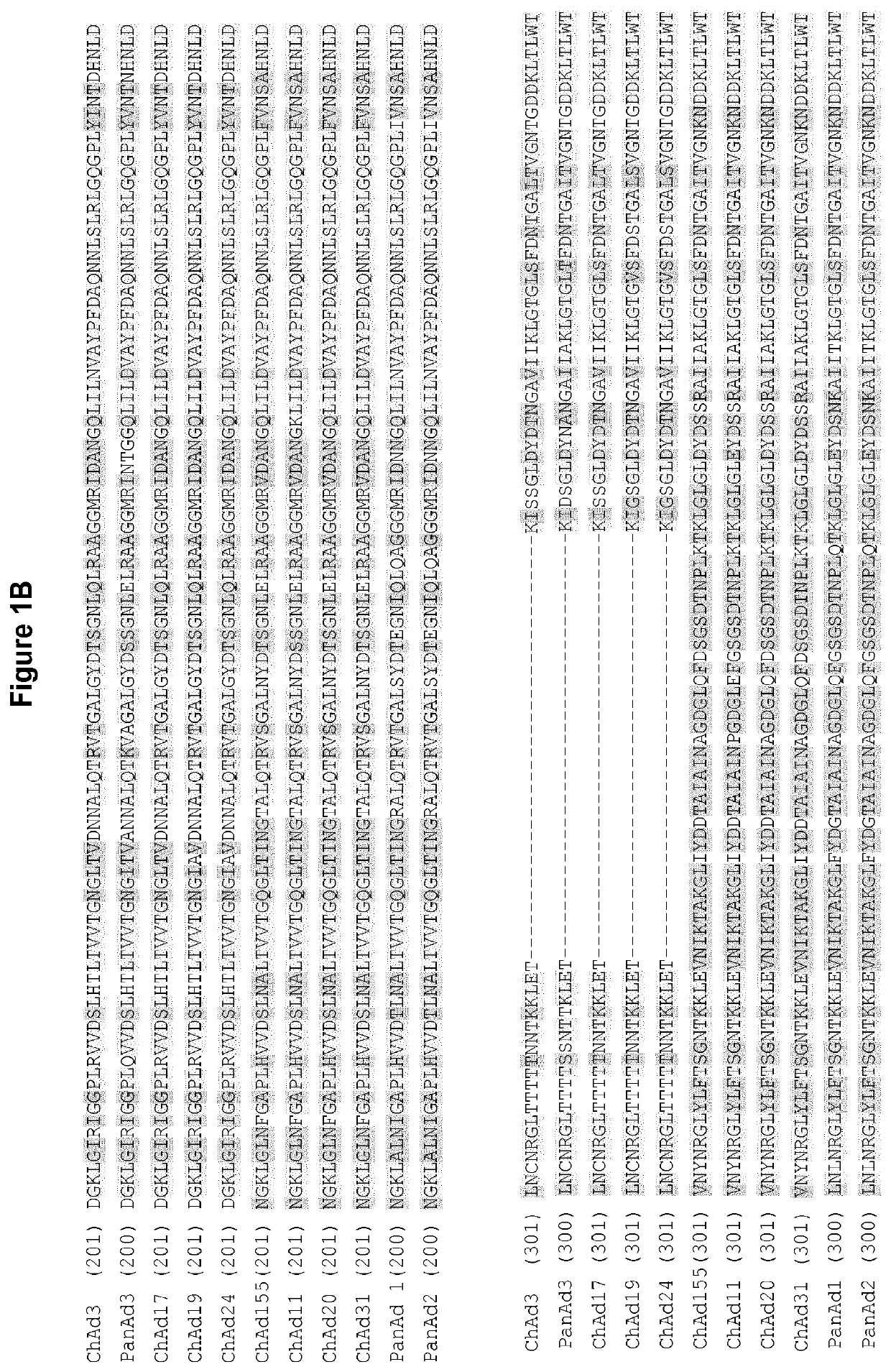
[0418]The ChAd155 viral genome was then cloned in a plasmid or in a BAC vector and subsequently modified (FIG. 2) to carry the following modifications in different regions of the ChAd155 viral genome:
[0419]a) deletion of the E1 region (from bp 449 to bp 3529) of the viral genome;
[0420]b) deletion of the E4 region (from bp 34731 to bp 37449) of the viral genome;
[0421]c) insertion of the F4orf6 derived from human Ad5.
[0422]2.1: Deletion of E1 Region: Construction of BAC / ChAd155 ΔE1_TetO hCMV RpsL-Kana #1375
[0423]The ChAd155 viral genome was cloned into a BAC vector by homologous recombination in E. coli strain BJ5183 electroporation competent cells (Stratagene catalog no. 2000154) co-transformed with ChAd155 viral DNA and Subgroup C BAC Shuttle (#1365). As shown in the schematic of FIG. 3, the Subgroup C Shuttle is a BAC vector derived from pBeloBAC11 (GenBank U51113, NEB) and which is dedicated to the cloning of ChAd belonging to species C and therefore contains the...
example 3
oduction
[0452]The productivity of ChAd155 was evaluated in comparison to ChAd3 and PanAd3 in the Procell 92 cell line.
[0453]3.1: Production of Vectors Comprising an HIV Gag Transgene
[0454]Vectors expressing the HIV Gag protein were prepared as described above (ChAd155 / GAG) or previously (ChAd3 / GAG Colloca et al, Sci. Transl. Med. (2012) 4:115ra). ChAd3 / GAG and ChAd155 / GAG were rescued and amplified in Procell 92 unto passages 3 (P3); P3 lysates were used to infect 2 T75 flasks of Procell 92 cultivated in monolayer with each vector. A multiplicity of infection (MOI) of 100 vp / cell was used for both infection experiments. The infected cells were harvested when full CPE was evident (72 hours post-infection) and pooled; the viruses were released from the infected cells by 3 cycles of freeze / thaw (−70° / 37° C.) then the lysate was clarified by centrifugation. The clarified lysates were quantified by Quantitative PCR Analysis with primers and probe complementary to the CMV promoter region....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com