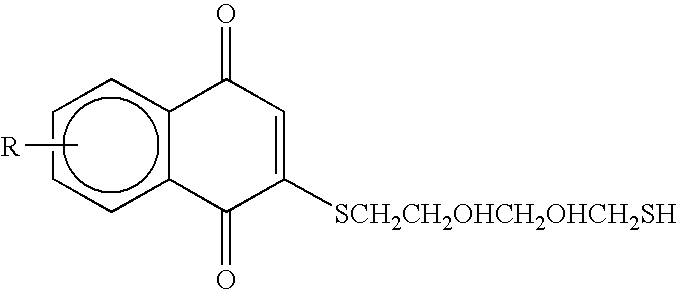
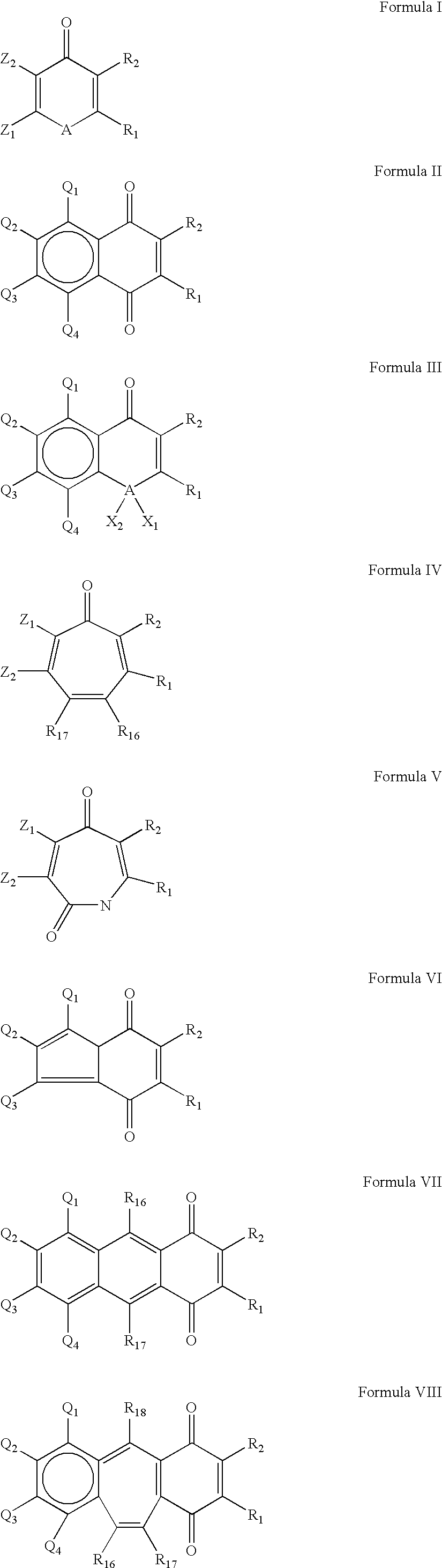
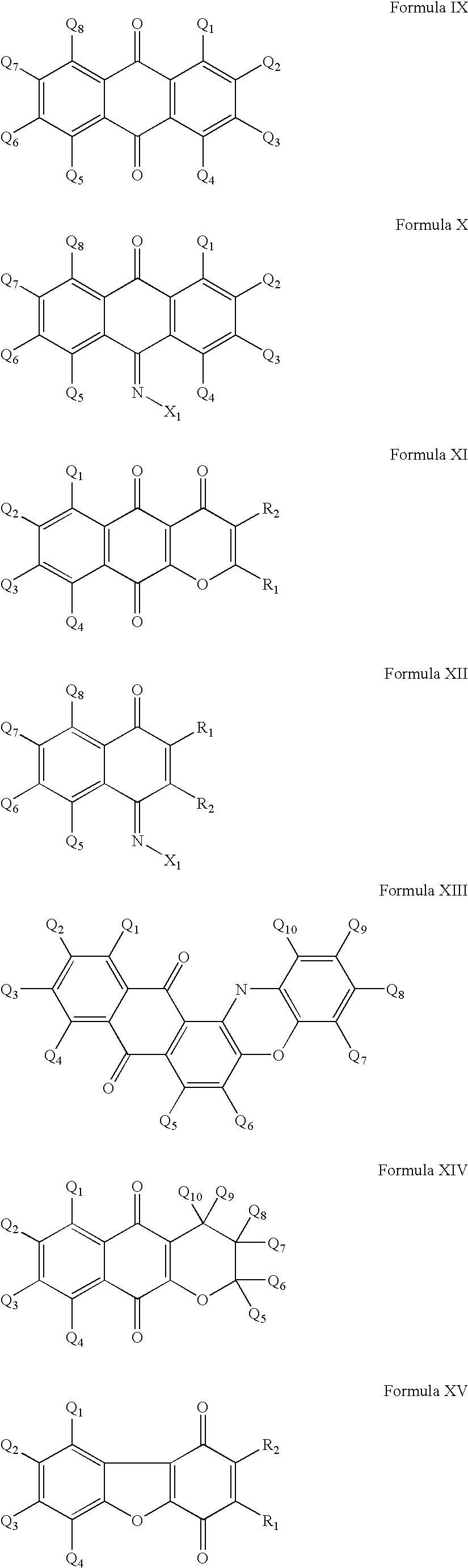
Cysteine protease inhimbitors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 2
Caspase-3 Inhibitors
[0091] Representative compounds of the present invention were purchased as part of a combinatorial library from Nanoscale Combinatorial Synthesis, Inc. (NANOSYN.RTM.; Mountain View, Calif.). A number of other compounds were purchased individually from commercial sources (i.e., Compound TestNumbers cpi0116-cpi0135). A few compounds were custom synthesized (i.e., Compound Test Numbers cpi0139-cpi0141).
[0092] To determine the caspase inhibitory activity of these compounds, the in vitro high throughput caspase-3 assay presented herein was utilized. Purified human recombinant caspase-3, fluorescence labeled substrate (Acetyl-Asp-Glu-Val-Asp-7-amino-4-trifluoromethyl coumarin), and known inhibitor (Z-Asp-Glu-Val-Asp-fluoromethyl ketone) were purchased from Sigma. The enzyme reactions were carried out at room temperature in 10 mM PIPES pH=7.4, 2 mM EDTA, 0.1% CHAPS, 5 mM DTT reaction buffer. Each well on the 96 well plate contained the above reaction buffer plus 55 .mu....
example 3
Computational QSAR Prediction of Blood-Brain Permeability
[0095] To assess whether the compounds of the present invention might be useful in treating neurodegenerative diseases, a QSAR (Quantitative Structure-Activity Relationship) model was used to computationally predict the blood-brain barrier permeability for each compound. We developed the QSAR model using the MOE software package from the Chemical Computing Group. A set of 75 compounds with known blood-brain partition coefficients were obtained from the literature (Luco, J. M. 1999. J Chem Inf Comput Sci 39:396-404). A set of 15 descriptors available in this software package were chosen based on a principle component analysis. The QSAR equation was then obtained by linear regression. The resulting QSAR prediction equation reproduced the test set log BB data to an accuracy of RMSE=0.375975 and R.sup.232 0.781358.
[0096] The results of the study are presented in Table IV as log BB values, i.e., the base ten log of the ratio of con...
example 4
Cross-Reactivity
[0097] To evaluate whether inhibition was specific to caspase-3 or applicable to other cysteine proteases or other proteases, inhibitor molecules were assayed for their capacity to inhibit additional non-caspase proteases. Three proteases, TPCK-trypsin, .alpha.-chymotrypsin and papain, were used to test for protease inhibition cross-reactivity. Protease inhibition assays were performed in 96-well flat-bottomed micro-titer plates with the QuantiCleave.TM. protease assay kit (Pierce, Rockford, Ill.) according to the manufacturer's instructions. Briefly, the assay conditions contained 100 .mu.M compound, 2 mg / mL of succinylated-casein and 0.15 mg / mL of the protease. The assay incubated in a final volume of 150 .mu.Ls 0.05 M sodium borate pH 8.5 buffer (NaB-buffer) for 20 minutes at 25.degree. C. After the protease reaction, 50 .mu.Ls of a trinitrobenzenesulfonic acid (TNBSA) solution (1:15, TNBSA:NaB-buffer) was added to each reaction and further incubated for 20 minute...
PUM
| Property | Measurement | Unit |
|---|---|---|
| humidity | aaaaa | aaaaa |
| humidity | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com