Nanoparticles for resisting H1N1 subtype influenza A virus based on self-assembled ferritin as well as preparation method and application of nanoparticles
An influenza virus and nanoparticle technology, applied in the field of genetic engineering, can solve the problem that antibodies cannot provide cross-protection, achieve high sensitivity and curative effect, improve accuracy and repeatability, and shorten the treatment process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Example 1 Construction of plasmid signal peptide-SpyTag-ferritin monomer subunit (signal peptide-ST-FE) and signal peptide-SpyCatcher-anti-H1N1 subtype influenza virus hemagglutinin protein Nanobody (signal peptide-SC-HA -VHH)
[0049] First, the gene signal peptide-ST-FE and the signal peptide-SC-HA-VHH were synthesized. The gene tandem structure is as follows figure 1 , and optimized by the codons of the CHO eukaryotic expression system, synthesized by GenScript Biotechnology Co., Ltd. The amino acid sequence of the signal peptide-ST-FE (SEQ ID NO: 1) is as follows:
[0050] MGWSCIILFLVATATGVHSAHIVMVDAYKPTKGGGSGGGSGGGSRMLKALNDQLNRELYSAYLYFAMAAYFEDLGLEGFANWMKAQAEEEIGHALRFYNYIYDRNGRVELDEIPKPPKEWESPLKAFEAAYEHEKFISKSIYELAALAEEEKDYSTRAFLEWFINEQVEEEASVKKILDKLKFAKDSPQILFMLDKELSARAPKLPGLLMQGGE;
[0051] The amino acid sequence (SEQ ID NO: 2) of the signal peptide-SC-HA-VHH is as follows:
[0052] MGWSCIILFLVATATGVHSSYYHHHHHHDYDIPTTENLYFQGSATHIKFSKRDEDGKELAGATMELRDSSGKTISTW...
Embodiment 2
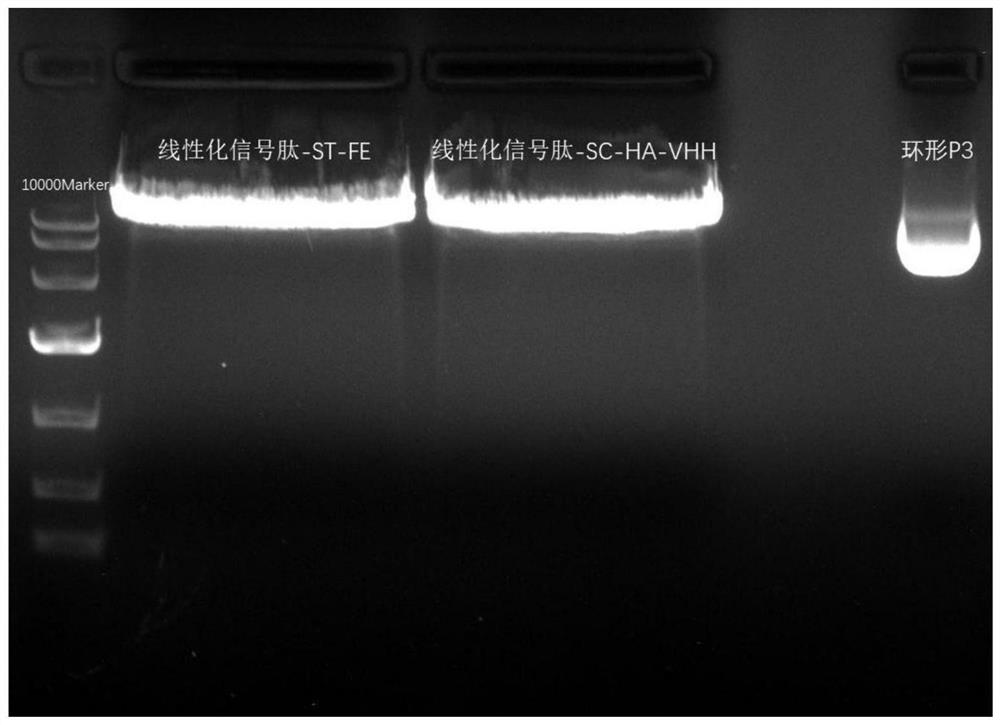
[0081] Example 2 Linearization of Plasmid Signal Peptide-ST-FE and Signal Peptide-SC-HA-VHH
[0082] Take out the plasmid signal peptide-ST-FE and signal peptide-SC-HA-VHH stored at -20°C, and measure their concentrations by NanoDrop one to be 1200ng / μL and 890ng / μL, respectively, and use the restriction endonuclease PvuⅠ for single enzyme Cut, concrete reaction system is shown in following table 3, table 4:
[0083] Table 3 Plasmid signal peptide-ST-FE single digestion reaction system
[0084]
[0085] Table 4 Plasmid signal peptide-SC-HA-VHH single enzyme digestion reaction system
[0086]
[0087] Add each component according to the single enzyme digestion reaction system table, and place it in a 37°C water bath for 2h digestion.
[0088] During digestion, prepare a 1% agarose gel. First, prepare the gel plate, select and insert the comb according to the total enzyme digestion system, then weigh 0.5g of agarose into a 100mL beaker, weigh 50mL of 1×TAE with a gradua...
Embodiment 3
[0089] Example 3 Electrotransfection of linear signal peptide-ST-FE and signal peptide-SC-HA-VHH GS gene deletion CHO cells
[0090] (1) Preparation before electroporation of CHO suspension cells lacking GS gene. 24h before electroporation, cells were at a viable cell density of 5×10 5 The cells / mL were inoculated into a 125 mL conical flask for shaking culture, the culture volume was 30 mL, and the rotational speed of the carbon dioxide shaking incubator was set to 110 rpm.
[0091] (2) Collection of CHO suspension cells lacking GS gene. On the day of electroporation, samples were taken to calculate the viable cell density and viability (98%) of suspension culture, and 1×10 cells were collected by calculation. 7 The cell suspension volume required for each viable cell was placed in a sterile centrifuge tube, centrifuged at 1000 rpm for 5 min, and then resuspended in 400 μL of 302 medium (containing glutamine).
[0092] (3) Incubation of cells with plasmids. Add 40 μg of l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com