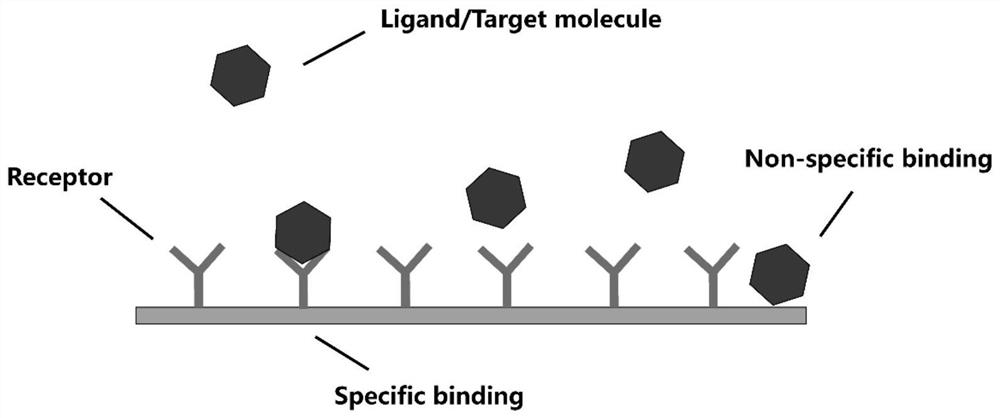
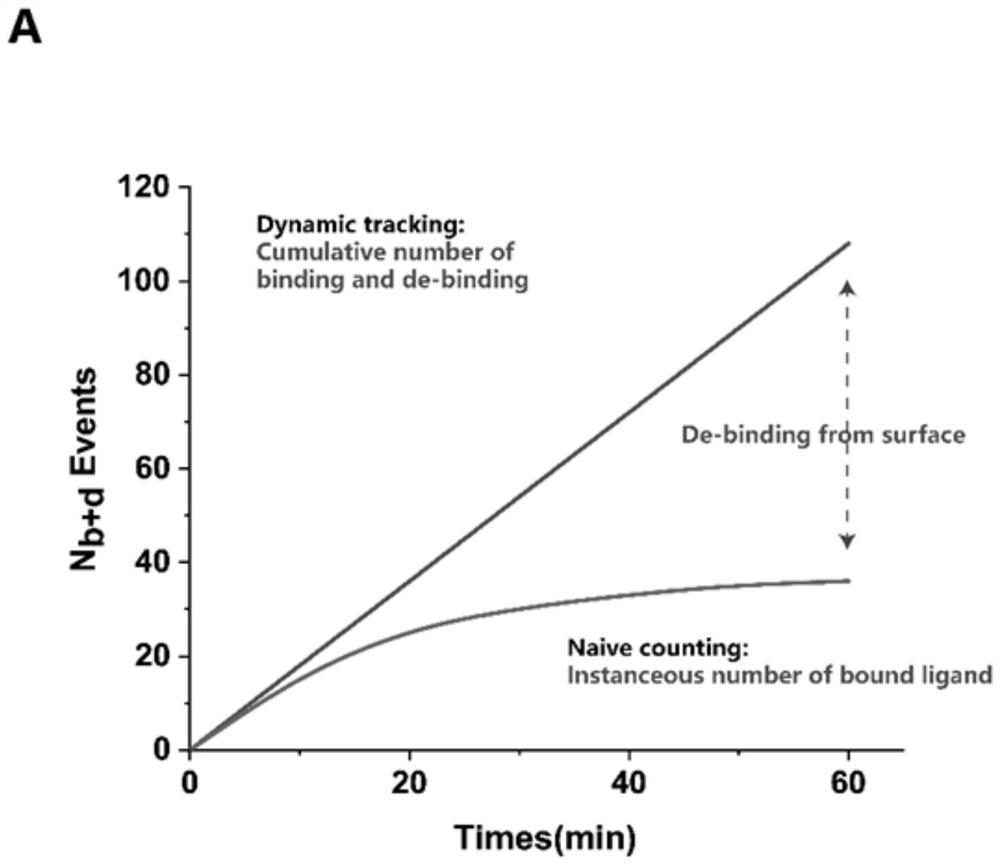
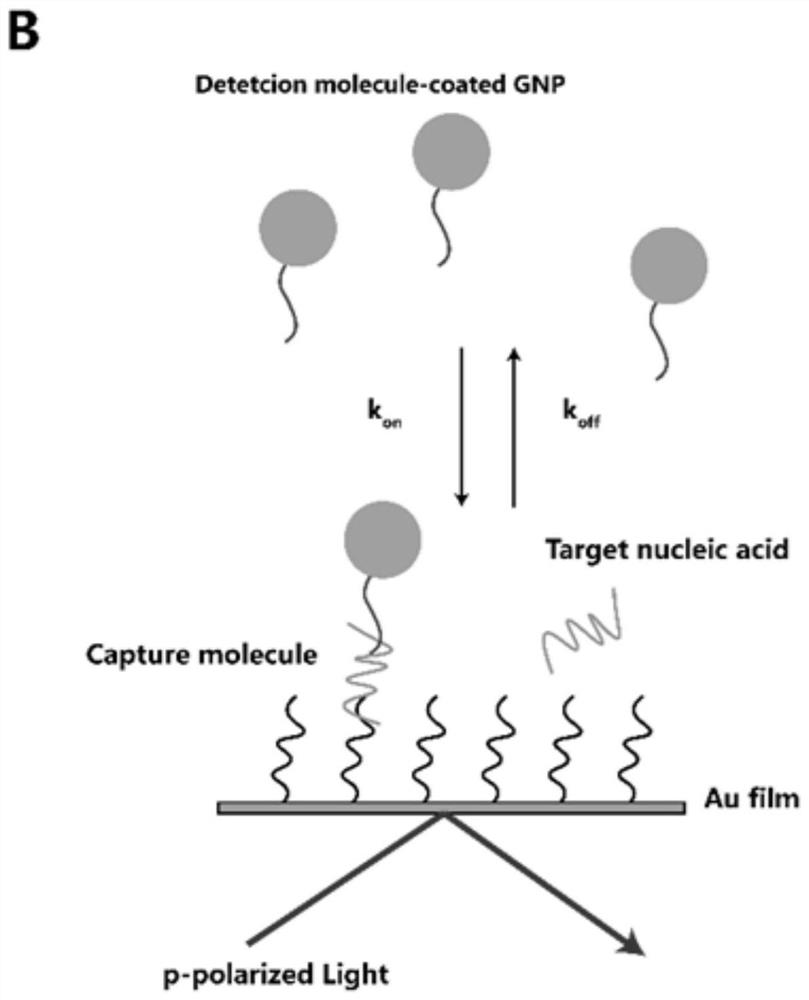
Amplification-free nucleic acid detection method
A technology for amplifying nucleic acid and detection methods, which is applied in the field of amplification-free nucleic acid detection, can solve problems such as aerosol pollution, inability to solve specificity, and inability to stabilize detection signals, so as to improve efficiency, achieve super-specificity, and improve sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] The base complementary chain length of the target nucleic acid and the detection molecule is 12base pair (the following detection 12bp) The non-amplification detection method of the target nucleic acid is as follows:
[0079] (1) Use a SPRM rebuilt based on a commercial total internal reflection microscope (Olympus IX-81) as a detection instrument, select an oil lens with a magnification of 100 times, and a numerical aperture N.A=1.49. The light source is an ultra-broadband light source SLED, and the light source controls the intensity of the current At 140mA, the field of view is 512×512pixels (full field of view: 33.28×33.28μm 2 ). A micro manager was used to control the CCD camera (Photometrics) to record the imaging signal of SPRi. Olympus microscope comes with Cell lens software to control the incident angle of the light path.
[0080] (2) On the BK-7 glass slide plated with 3nm chromium and 47nm gold, first wash and dry it with piranha lotion, and then treat it ...
Embodiment 2
[0085] The non-amplification detection method of the target nucleic acid with a base complementary chain length of 11 bp between the target nucleic acid and the detection molecule is as follows:
[0086] (1) Use a SPRM rebuilt based on a commercial total internal reflection microscope (Olympus IX-81) as a detection instrument, select an oil lens with a magnification of 60 times and a numerical aperture N.A=1.49, and the light source is an ultra-broadband light source SLED, and the light source controls the intensity of the current At 150mA, the field of view is 512×512pixels(full field of view:51.2×51.2μm 2 ). A micro manager was used to control the CCD camera (Photometrics) to record the imaging signal of SPRi. Cell lens software is used to control the incident angle of the light path.
[0087] (2) On the BK-7 glass slide coated with 2nm chromium and 47nm gold, first wash and dry it with alcohol and pure water, and then treat it with hydrogen flame to remove surface particl...
Embodiment 3
[0092] The non-amplification detection method of the target nucleic acid with a base complementary chain length of 10 bp between the target nucleic acid and the detection molecule is as follows. In this example, the detection molecule is immobilized on the chip surface, and the capture molecule is combined with the nanoparticle:
[0093] (1) Use a SPRM rebuilt based on a commercial total internal reflection microscope (Olympus IX-81) as a detection instrument, select an oil lens with a magnification of 60 times and a numerical aperture N.A=1.49, and the light source is an ultra-broadband light source SLED, and the light source controls the intensity of the current At 169mA, the field of view is 512×512pixels(full field of view:51.2×51.2μm 2 ). A micro manager was used to control the CCD camera (Photometrics) to record the imaging signal of SPRi. Cell lens software is used to control the incident angle of the light path.
[0094] (2) On the BAK-4 glass slide coated with 2nm c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com