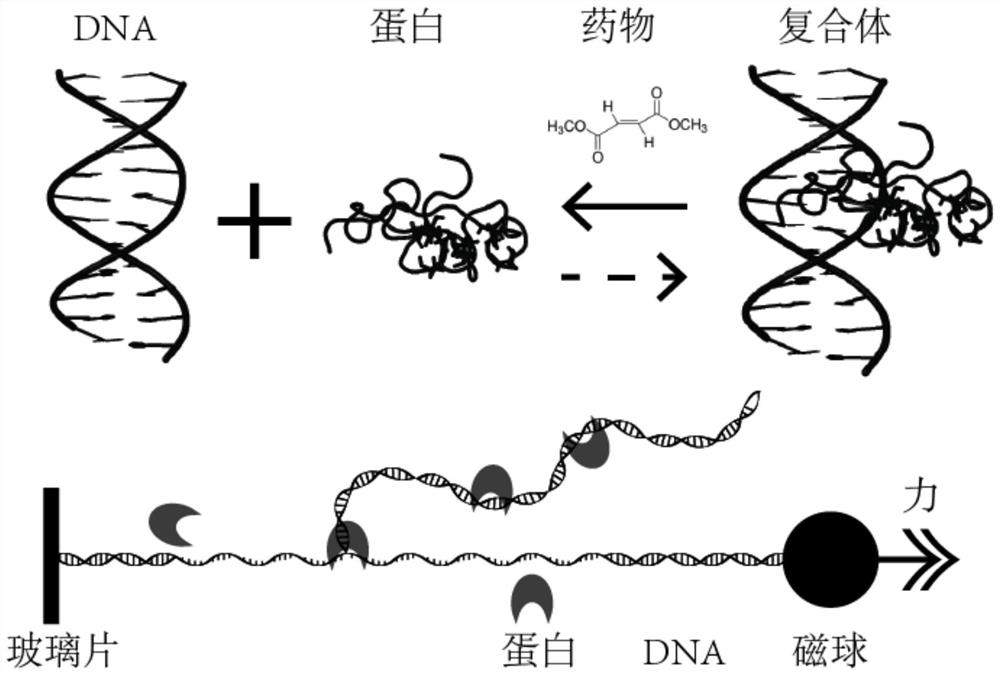
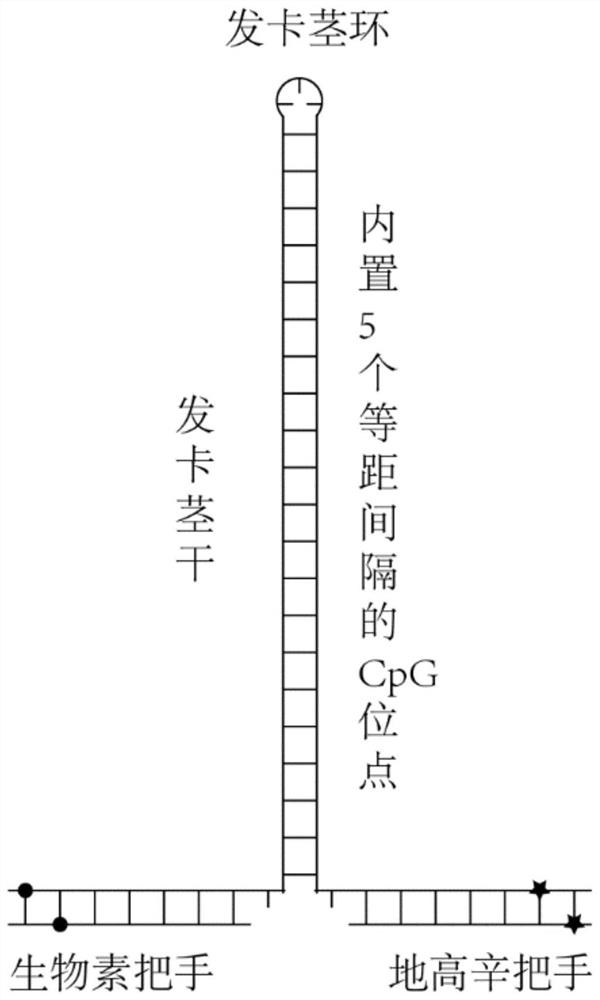
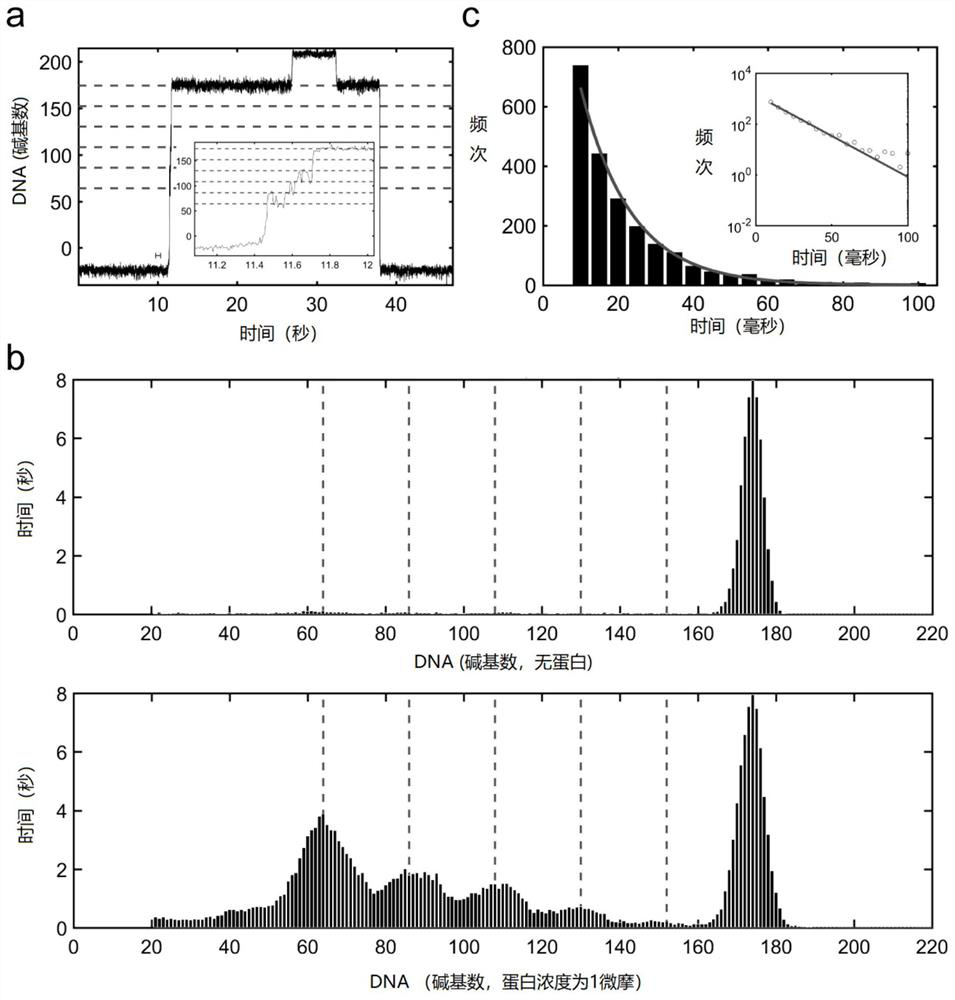
Single-molecule mechanical method for measuring inhibition of small-molecule drug on protein and nucleic acid interaction
A technology for inhibiting proteins and small molecules, which is applied in measuring devices, scientific instruments, and analytical materials, etc., and can solve the problems of single-molecule mechanics measurement of small-molecule drug inhibitory protein-nucleic acid interaction methods that have not been reported.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 Constructing a DNA structure-based unimolecular reactor for CpG motifs
[0048] Table 1 Artificial oligonucleotide DNA name and sequence information
[0049]
[0050]
[0051]DNA hairpin construct handle preparation. The handle of the DNA hairpin structure contains biotin and digoxin modification, and is prepared by polymerase chain reaction, and the process is as follows. Add 1 µl of biotin / digoxigenin handle upstream primer (Table 1, concentration 10 µM), 1 µl biotin / digoxigenin handle downstream primer (Table 1, concentration 10 µM) to an enzyme-free reaction tube 10 micromolar, primers were purchased from Sangon Bioengineering Company), 1 microliter (concentration: 10 millimolar) of dATP (article number: 4026Q, Baoriji Biotechnology Co., Ltd.), 1 microliter (concentration: 10 millimolar) of dATP dGTP (Cat. No.: 4027Q, Biotech Biotechnology Co., Ltd.), 1 microliter of dCTP (concentration: 10 millimolar, Cat. No.: 4028Q, Biotech Biotech Co., Ltd.), 0...
Embodiment 2
[0056] Example 2 Establishment of a single-molecule reactor system based on a DNA hairpin structure in a microfluidic reaction pool
[0057] The preparation process of the microfluidic reaction cell is as follows. Take two slides, the size is 60 mm × 24 mm. On a multi-functional grinding and engraving machine (model: TC-169), punch holes at both ends of a glass slide for liquid sampling and sampling.
[0058] Then, perform a washing step. Slides were immersed in water with dish soap and ultrasonically cleaned for 30 minutes. Replace with pure water and wash 3 times, 5 minutes each time. Replace the isopropanol and sonicate for 30 min. Replace pure water, wash 3 times, 5 minutes each time. Replace with 75% ethanol and soak for several hours. Finally blow dry with nitrogen.
[0059] Spread the nitrocellulose and fix the reference pellet. Take 1% nitrocellulose stock solution, add 250mg / 10ml monodisperse polypropylene beads (article number: PSC-03000, Big Goose (Tianjin) ...
Embodiment 3
[0064] Example 3 Normalization processing of unimolecular reaction data lines of MLL1 CXXC protein binding to CpG motif
[0065] The protein used in this example is the MLL1 CXXC domain, which can specifically recognize the CpG motif contained in the stem of the DNA hairpin structure. MLL1 CXXC protein was synthesized by Sangon Bioengineering Company. In this embodiment, when detecting the binding position of the protein on the DNA, the length of the DNA is first normalized according to the base unit. Under the test force conditions, the normalization process is based on the length of the DNA hairpin structure after it is completely unzipped. Each single-molecule data line is self-calibrated according to the length of the DNA after melting, and then a histogram is built. The middle peak in the histogram distribution indicates where the protein binds to the DNA. Convert DNA length from nanometers to bases with reference to the middle peak of the histogram distribution. This...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com