Broad-spectrum proteome editing with an engineered bacterial ubiquitin ligase mimic
A protein and bacterial technology, applied in the field of broad-spectrum proteome editing, can solve the problem of low efficacy of PROTAC
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0236] The following examples are provided to illustrate embodiments of the application, but they are in no way intended to limit the scope thereof.
[0237] The following examples are included to illustrate illustrative embodiments of the present application. It should be appreciated by those of skill in the art that the techniques disclosed in the examples which follow represent techniques discovered by the inventors to function well in the practice of this application, and thus can be considered to constitute preferred modes for its practice. However, those of skill in the art should, in light of the present application, appreciate that many changes can be made in the specific embodiments which are disclosed and still obtain a like or a similar result without departing from the spirit and scope of the application.
[0238] experimental method
[0239] plasmid. Table 1 provides all plasmids used in this study.
[0240] Table 1. Strains, cell lines and plasmids used in thi...
example 1
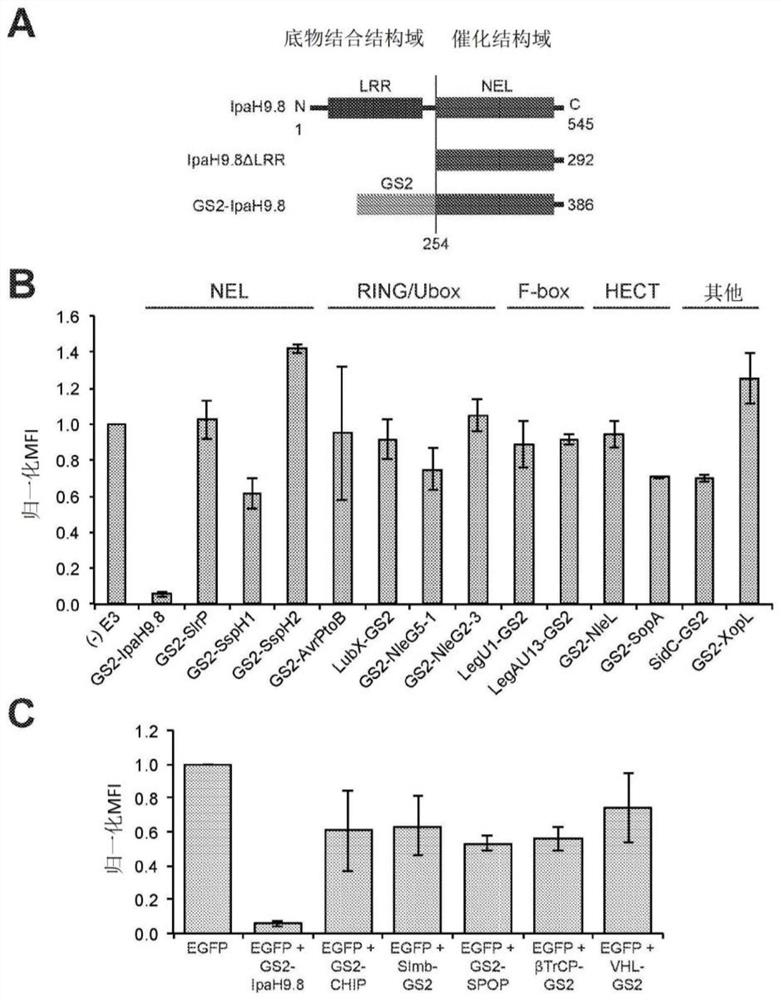
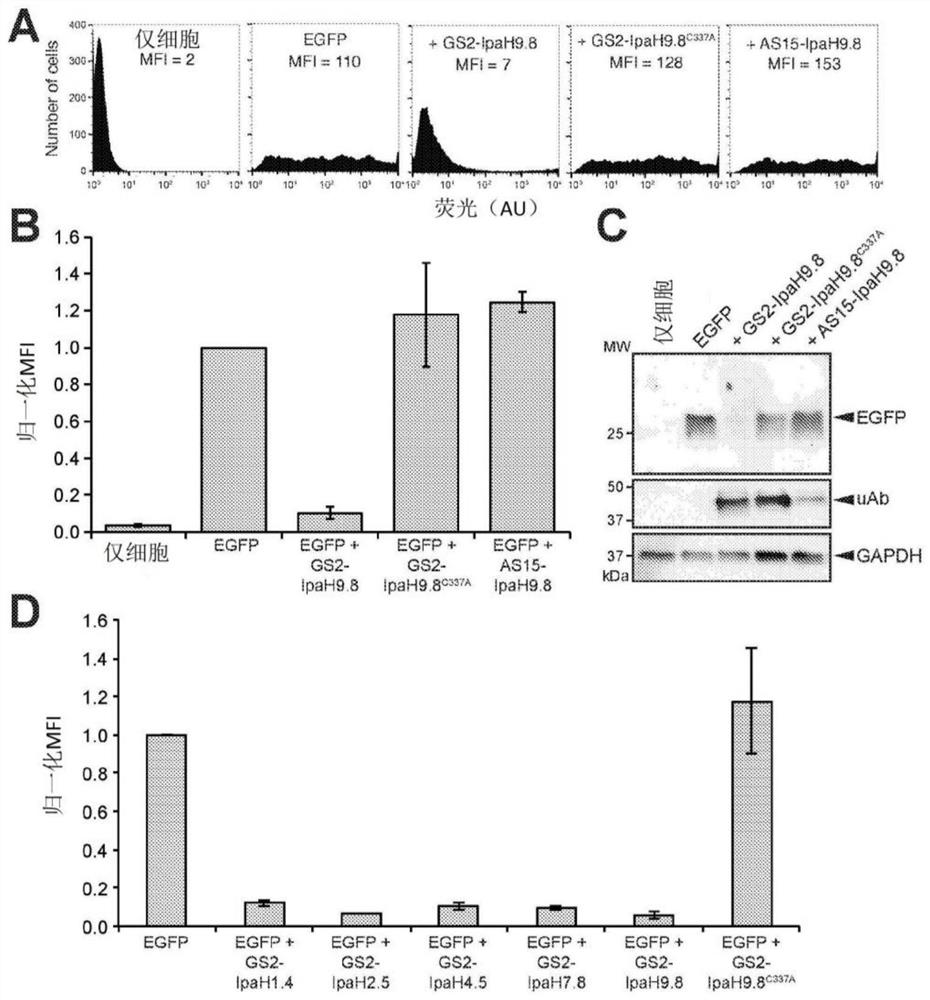
[0275] Example 1 - Engineered IpaH9.8 efficiently silences GFP in mammalian cells
[0276] To determine whether E3 ubiquitin ligase mimics from pathogenic bacteria could be redesigned to silence non-native targets, we focused on a panel of 14 candidate enzymes that represent the major E3 classes found in bacteria to date (Table 1, below). 2). Maculins et al., “Bacteria-Host Relationship: Ubiquitin Ligases as Weapons of Invasion. Cell Res. 26(4):499-5l0 (2016) and Lin et al., “Exploitation of the Host Cell Ubiquitin Machinery by Microbial Effector Proteins,” J. Cell Sci. 130(12):1985-96(2017), the entire contents of which are hereby incorporated by reference.
[0277] Table 2. Bacterial E3 ubiquitin ligases evaluated in this study.
[0278]
[0279]
[0280] 1 References listed in Table 2 provide clear evidence or annotations of the catalytic domain of each E3 ubiquitin ligase.
[0281] Abbreviations in Table 2: NEL, novel E3 ligase; HECT, homology to E6AP C-terminus;...
example 3
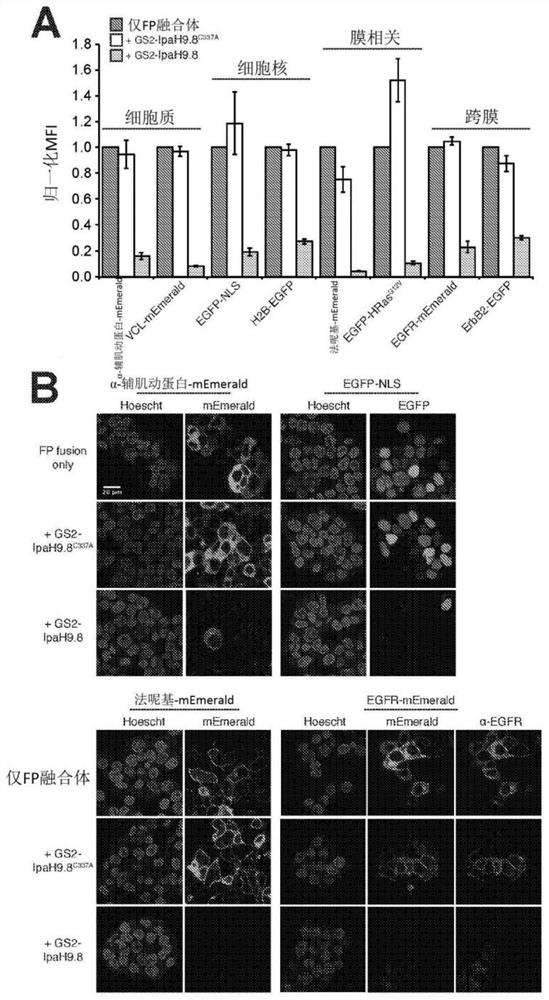
[0305] Example 3 - GS2-IpaH9.8-mediated proteome editing is flexible and modular.
[0306] An attractive feature of uAbs is their highly modular structure—the E3 catalytic domain and synthetic binding protein domain can be interchanged to reprogram activity and specificity. Indeed, the above results reveal the ease with which different bacterial and eukaryotic E3 domains can be chimerized to form functional uAbs. To investigate the interchangeability of synthetic binding protein domains in IpaH9.8-based uAbs, other high-affinity GFP-binding proteins such as the FN3 antibody analog GS5 (K d = 62nM) (Koide et al., "Teaching an Old Scaffold New Tricks: Monobodies Constructed Using Alternative Surfaces of the FN3 Scaffold," J Mol. Biol. 415(2):393-405 (2012), the entire contents of which are hereby incorporated by reference entered this article) or cAbGFP4 (K d = 0.32nM) to replace GS2 (Saerens et al., "Identification of a Universal VHH Framework to Graft Non-Canonical Antigen-B...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com