Multiple primers, kit and method for high-throughput sequencing of enterovirus
A technology of enteroviruses and kits, applied in the analysis of high-throughput sequencing data, and in the field of multiple primers, can solve the problems of systematic monitoring, early warning and prevention and control of enteroviruses, and the study of the rules of virus molecular variation, etc. problem, to achieve rapid identification, perfect detection and analysis technology, and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0095] Embodiment 1 multiplex PCR primer design
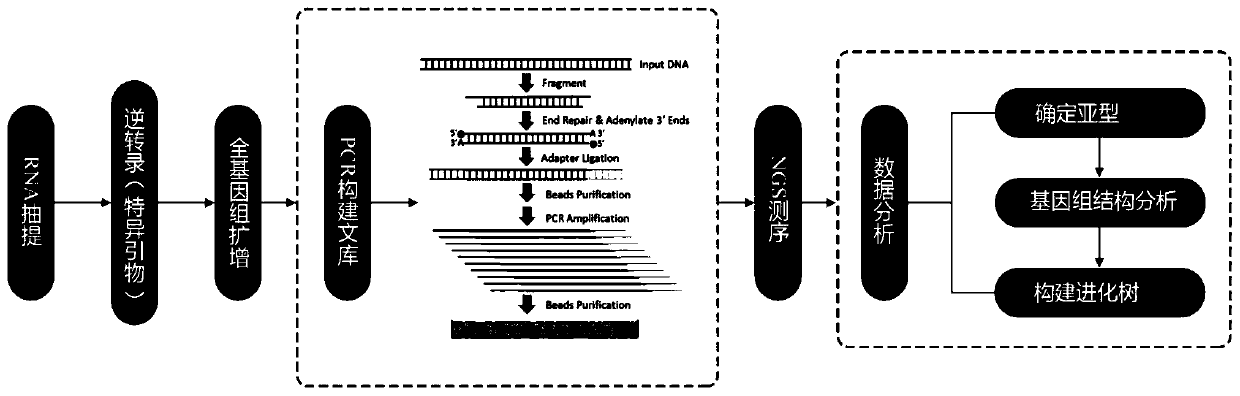
[0096] The base sequences of various genotypes of enteroviruses were obtained by using the public database (NCBI) and the resources of the Guangdong Province bacterial and virus strain bank, and the conserved segments at the head, tail and middle of the genome were identified through bioinformatics analysis. Different from the previous method of designing multiple primer sets to complete genome amplification through first-generation sequencing, and also different from the recently published method of determining the viral genome through polyA enrichment combined with next-generation sequencing, this example focuses on enterovirus A / B / C Conserved segment of the three genomes, divide the whole enterovirus genome into two segments of 4kb and 3.5kb, and design universal primers for the three groups of human enterovirus A / B / C in the conserved segment, each group contains two For primers, a total of six pairs of primers cover the en...
Embodiment 2
[0097] Embodiment 2 kit preparation
[0098] An embodiment of the kit that can be used for the construction and detection of the whole genome library of enteroviruses in the present invention, which includes reverse transcription reagents (reverse transcription primers, reverse transcription reaction solution, reverse transcriptase mixture, dNTP), multiplex PCR amplification Reagents (multiple PCR reaction solution, multiple PCR enzyme, multiple PCR primer mix 1, multiple PCR primer mix 2), library construction reagents (end repair reaction solution, end repair enzyme, ligation buffer, DNA ligase, linker, Specific adapters 1-12, library amplification reaction solution, library amplification enzyme). Wherein, multiplex PCR primer mixture 1 is formed by mixing polynucleotides SEQ ID NO.1 and 2, SEQ ID NO.1 and 5, SEQ ID NO.1 and 8; multiplex PCR primer mixture 2 is composed of polynucleotide SEQ ID NO.3 and 4, SEQ ID NO.6 and 7, and SEQ ID NO.9 and 10 are mixed.
[0099] Accor...
Embodiment 32019
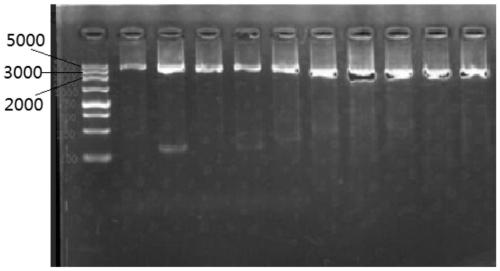
[0115] Example 3 Detection of Nosocomial Infection Epidemic Samples of Enterovirus in 2019
[0116] Compared with the current fluorescent quantitative PCR (RT-PCR) method, the present invention has the advantage of providing a universal sequencing method for the whole genome of enteroviruses based on a high-throughput detection platform, which overcomes the need to design specific primers for specific types in conventional RT-PCR Disadvantages of Probes. More importantly, the acquisition of the complete genome sequence of enteroviruses can not only accurately identify enterovirus types, but also further analyze the characteristics of the virus genome. In April 2019, a nosocomial infection of enterovirus broke out in Shunde, Guangdong Province, causing 4 newborn deaths and dozens of people showing symptoms of infection, causing serious social impact. Now take this epidemic as an example to illustrate how to use the kit in Example 2 to perform rapid sequence determination, typi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com