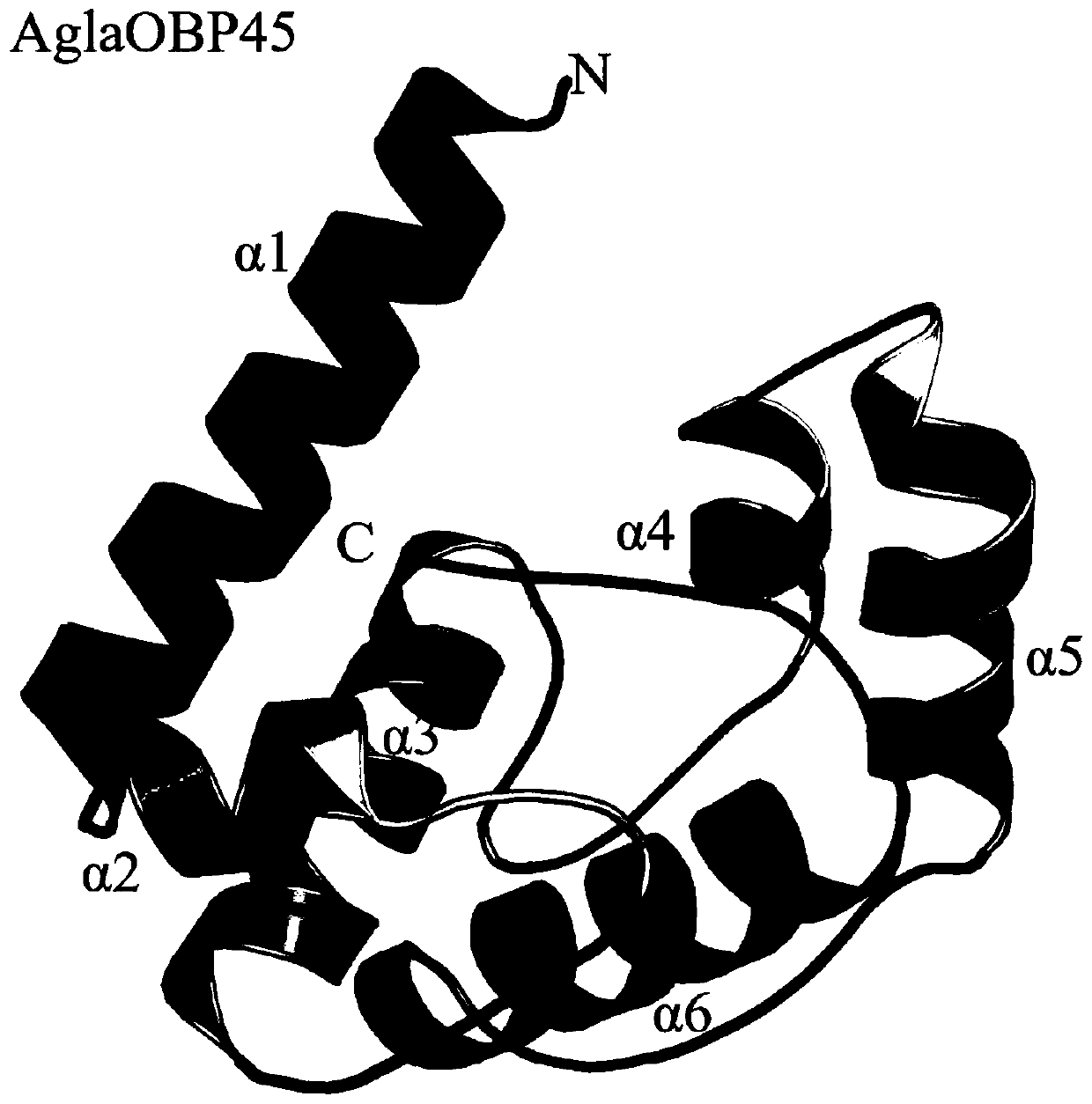
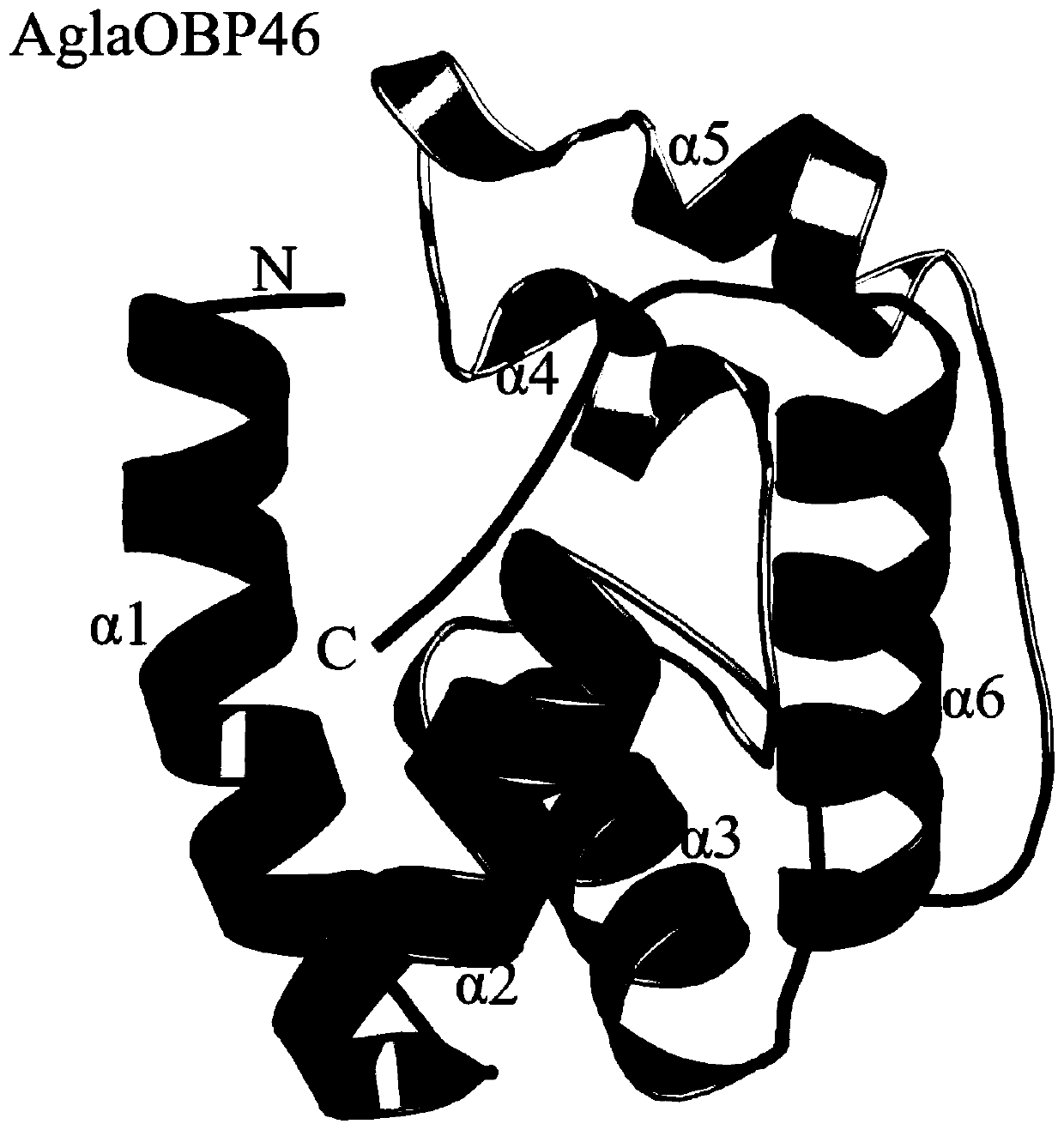
Anoplophora glabripennis odorant binding protein OBP45, OBP46 and application in screening attractants thereof
An odor-binding protein and A. chinensis technology, which can be applied in applications, proteomics, animal/human peptides, etc., can solve problems such as unsatisfactory attracting effect, and achieve the effect of improving screening efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1 Extraction and reverse transcription of total RNA from the antennae of Beetle glabrata
[0035] 1.1 Extraction of total RNA from antennae of Beetle glabrata
[0036] Collect the antennae of the newly emerged male and female Beetles glabrata, and extract the total RNA of the antennae, using TRIzol reagent (Ambion) and RNeasy Plus Mini kit (No.74134; Qiagen, Hilden, Germany) according to the instructions, and the specific steps are as follows:
[0037] (1) Take 50-100 mg of antennae of male and female adults of Beetleus glabrata and Beetleus glabrata respectively, put them in a 2ml centrifuge tube with grinding beads, add 1ml of Trizol reagent, shake and grind the cells;
[0038] (2) Place at room temperature for 30 minutes to reduce DNA contamination;
[0039] (3) 4°C, 12000g, 10min centrifugation;
[0040](4) Take about 800 μl of the supernatant into a clean 1.5ml centrifuge tube;
[0041] (5) Draw 3-5 times with a sterile syringe, cut genomic DNA and lyse ...
Embodiment 2
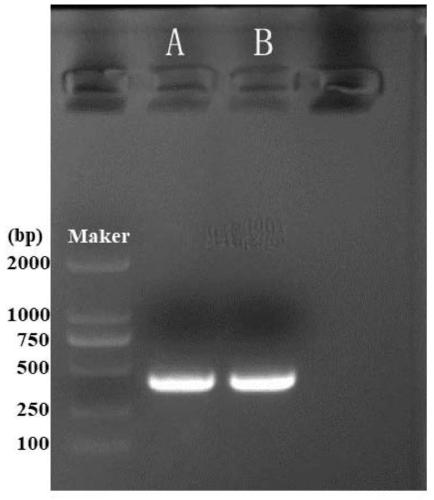
[0061] Cloning of Example 2 Odor Binding Protein OBP45 and OBP46 Genes of Beetle glabrata
[0062] 2.1 Primer design and PCR reaction
[0063] According to the sequences of AglaOBP45 and AglaOBP46 obtained from the transcriptome sequencing of the antennae of Beetle glabrata, the online software Primer3( http: / / bioinfo.ut.ee / primer3-0.4.0 / ) respectively designed the following primers to clone the entire ORF regions of AglaOBP45 and AglaOBP46.
[0064] AglaOBP45F: GGACAACTGCAACTCTTTGTCG
[0065] AglaOBP45R: GAGACCACAGATGGTGATGAGC
[0066] AglaOBP46F: GGTTCTGGTGATTGTGTATTTGG
[0067] AglaOBP46R: TACTCGCCGGTCCGTAAGATAG
[0068] Use TAKARA’s high-fidelity enzymes for PCR amplification of the target gene, configure the system on ice, SYBR PremixEx Taq II 12.5 μL, forward primer 0.75 μL, reverse primer 0.75 μL, template cDNA 1 μL, ddH 2 O 0.75 μL, total volume 25 μL. The PCR reaction conditions were: 98°C, 10s pre-denaturation; 55°C, 5s; 72°C, 5s, 34 cycles. The product was ...
Embodiment 3
[0080] The construction of embodiment 3 prokaryotic expression vector
[0081] 3.1 Amplification of target fragments and extraction of recombinant plasmids
[0082] Based on the cloned full-length gene sequences of AglaOBP45 and AglaOBP46, the signal peptide sequence was removed, and primers with enzyme cleavage sites (NdeI: CATATG, XhoI: CTCGAG) were designed to amplify the target fragment. The primer sequences are as follows:
[0083] AglaOBP45F:ACTTACATATGCTGATAAGATTAGGTGCCGCGTGC
[0084] AglaOBP45R: ACTACTCGAGTTATACTAAGAAGTAAGCATCTGGA
[0085] AglaOBP46F: ACTTACATATGCTGATAAGATTAGGTGCCGCGTGC
[0086] AglaOBP46R: ACTACTCGAGTTACGGATGGGGCAACTTGGGAA
[0087] After the cloned strains were sequenced and verified to be correct, they were expanded and cultivated. Then use the AxyPrep Plasmid DNA Mini Kit to extract the plasmid to obtain the plasmid with the target fragment.
[0088] 3.2 Double digestion of cloning plasmid and expression vector and connection of restriction pro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com