Sequencing library construction kit and using method and application thereof
A technology for sequencing libraries and kits, applied in the field of gene detection, can solve the problems of limited sites, contamination of PCR products, cumbersome operation process, etc., and achieve the effect of improving utilization efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] The invention provides a kit for constructing a sequencing library, specifically comprising a restriction endonuclease, a restriction endonuclease buffer, an end repair reagent, a DNA ligase reagent set, a ligase buffer, and a sequencing linker (01 -96), PCR primer set, PCR reaction system.
[0044] The restriction endonuclease is one or a combination of AseI, CviQI, NdeI, AciI, AluI, BfaI, HaeIII, HhaI, MseI, AsiSI, CviAII, PacI, PvuI, PvuI-HF, HpyCH4IV, HpySE526I, It should be understood that different restriction endonucleases can be selected according to the objects of the sequencing library, including but not limited to the restriction endonucleases listed above.
[0045] The end repair reagent group includes T4PNK (T4 Polynucleotide Kinase, T4 polynucleotide kinase), T4 DNA Polymerase (T4 DNA polymerase), Klenow DNA Polymerase (Klenow DNA polymerase), dNTPs, repair buffer.
[0046]The ligase in the DNA ligase reagent group can be DNA ligase, T4 DNA ligase, taq DN...
Embodiment 2
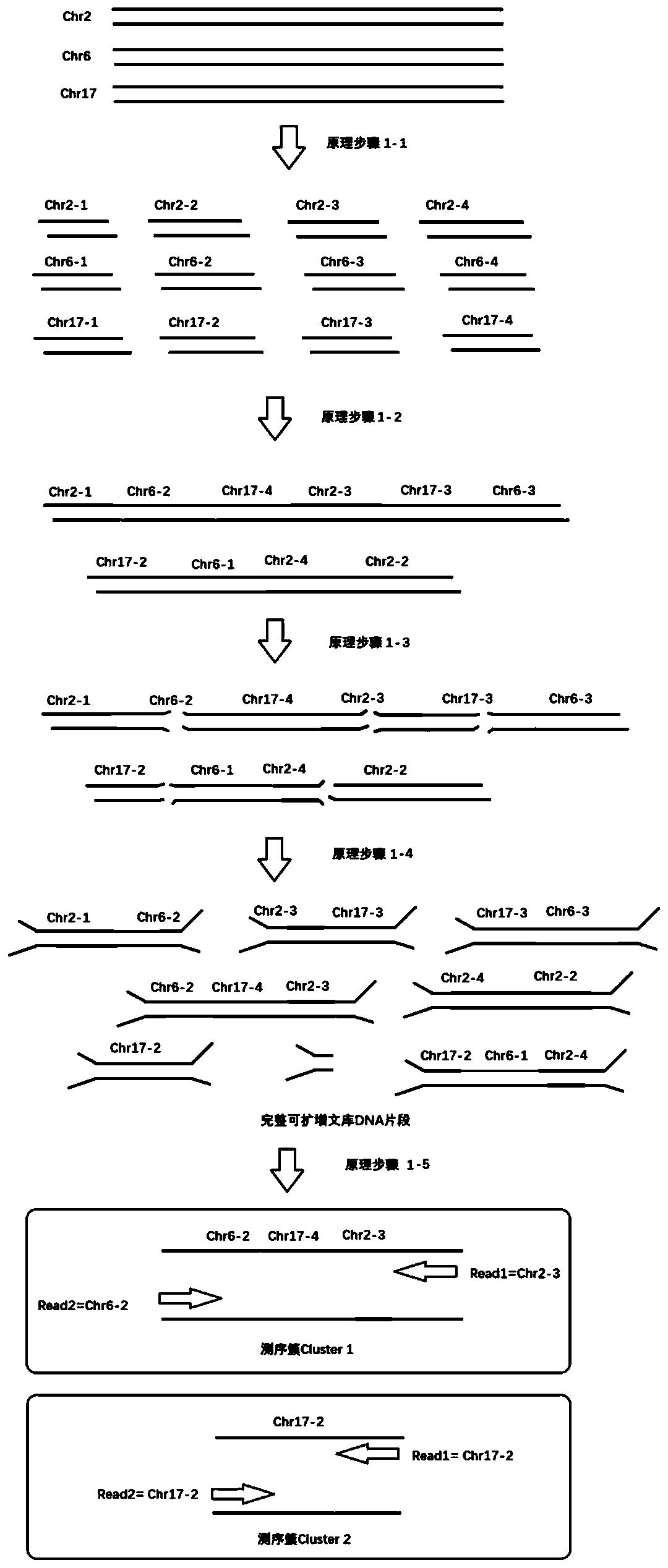
[0051] attached figure 1 A schematic diagram of a sequencing library construction method is shown, which is described in detail as follows:
[0052] Principle step 1-1: After the different chromosomes of the sample genome are cut by the restriction endonuclease reagent group, fragments of different sizes are formed, and these fragments all have 5'-phosphate groups and 3'-hydroxyl groups that can be reconnected ;
[0053] Principle steps 1-2: In the presence of the DNA ligase reagent group, different DNA fragments are randomly connected and become long fragments again;
[0054] Principle steps 1-3: Use random interruption methods such as ultrasonic interruption to randomly interrupt the re-formed long fragments again to obtain fragments from different regions;
[0055] Principle steps 1-4: After ligation with sequencing adapters, a complete library DNA fragment that can be amplified is formed;
[0056] Principle steps 1-5: During sequencing, there will be at least two differ...
Embodiment 3
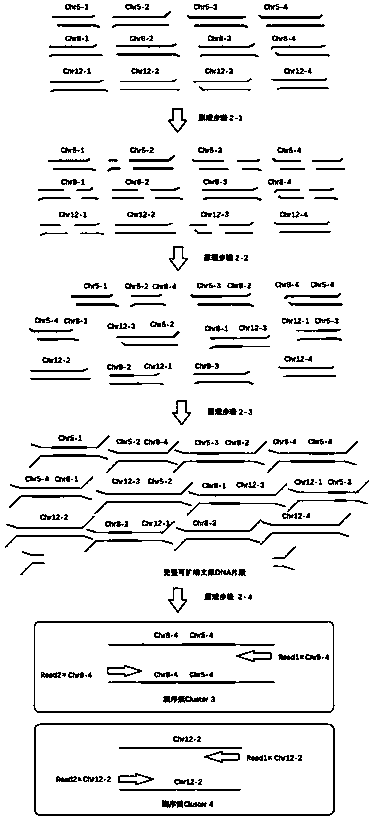
[0092] attached image 3 A schematic diagram of a sequencing library construction method is shown, which is described in detail as follows:
[0093]Principle step 2-1: The genomic DNA of the sample is first interrupted by ultrasound to form fragments of different sizes that need to be repaired at the ends and are not easy to connect;
[0094] Principle step 2-2: Add a restriction enzyme reagent group, the DNA fragment containing the endonuclease recognition site will be cut to form a port that can be connected with a 5'-phosphate group and a 3'-hydroxyl group;
[0095] Principle step 2-3: In the presence of DNA ligase reagent group, different DNA fragments with ports that can be connected are randomly connected to form a new hybrid fragment; no endonuclease recognition site, no restriction Fragments cut by endonucleases will not be ligated;
[0096] Principle steps 1-4: After end repair and ligation with sequencing adapters, complete library DNA fragments that can be amplifi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com