Method for detecting CLC gene expression quantity in nasal cavity exfoliated cells and application of method
A technology of gene expression and exfoliated cells, which is applied to the detection of CLC gene expression in nasal exfoliated cells, can solve the problems of non-healing mucosal pathological biopsy, failure to reflect the overall picture of the tissue, and increase the risk of infection in patients
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0059] The preparation method of the DNase reaction solution comprises the following steps: taking DNase buffer solution, recombinant DNase, and double distilled water without RNase and mixing to obtain the DNase reaction solution. Preferably, the preparation method of the DNase reaction solution comprises the following steps: take 5 μL of 10×DNase buffer solution, 4 μL of recombinant DNase, and 41 μL of RNase-removed double-distilled water and mix to obtain the DNase reaction solution.
[0060] As a preference, when the genome content is low or the initial amount of material is low when the RNA is extracted, the method for extracting RNA from nasal exfoliated cells further includes the following steps: in the step 1, the nasal exfoliated cells are lysed After being added to the cell lysate, it is first added to the genomic DNA adsorption column to obtain the filtrate, and then an equal volume of ethanol is added to the filtrate.
[0061] Preferably, in order to obtain a highe...
Embodiment 1
[0091] Sample collection and processing:
[0092] After rinsing the nasal cavity with normal saline, a patient pressed a brush (manufactured by Copan) on the surface of the nasal polyp for 30 seconds under the nasal endoscope, rotated it 3 to 4 times, and brushed the surface of the polyp. ℃ for short-term storage (not exceeding 24 hours), or transfer to below -20 ℃ for long-term storage.
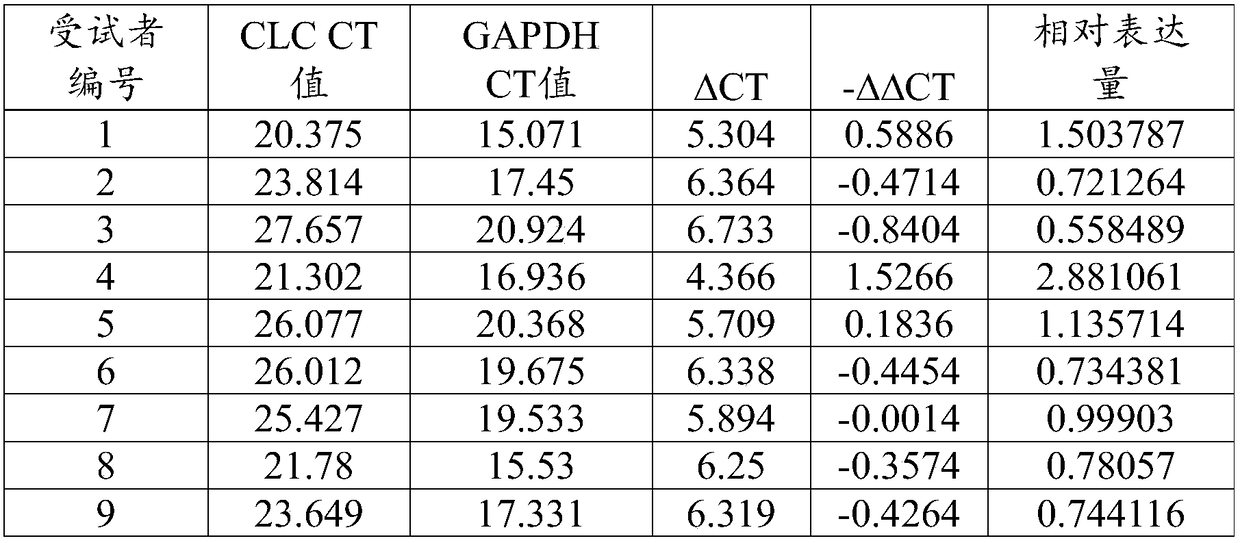
[0093] A method for detecting CLC gene expression in nasal cavity exfoliated cells, comprising the following steps:
[0094] Step 1: Extract RNA from nasal exfoliated cells:
[0095] Step 1: Take the genomic DNA adsorption column and place it in a 2mL collection tube, dissolve the nasal exfoliated cells in 300 μL of cell lysate and add them to the genomic DNA adsorption column, take the filtrate, and add an equal volume of 70% Ethanol, mixed evenly, was added to the RNA purification column, 12000 rpm, centrifuged for 1min, the filtrate was removed, and the RNA purification column was place...
Embodiment 2
[0106] Sample collection and processing:
[0107] After rinsing the nasal cavity with normal saline, a patient pressed a brush (manufactured by Copan) on the surface of the nasal polyp for 30 seconds under the nasal endoscope, rotated it 3 to 4 times, and brushed the surface of the polyp. ℃ for short-term storage (not exceeding 24 hours), or transfer to below -20 ℃ for long-term storage.
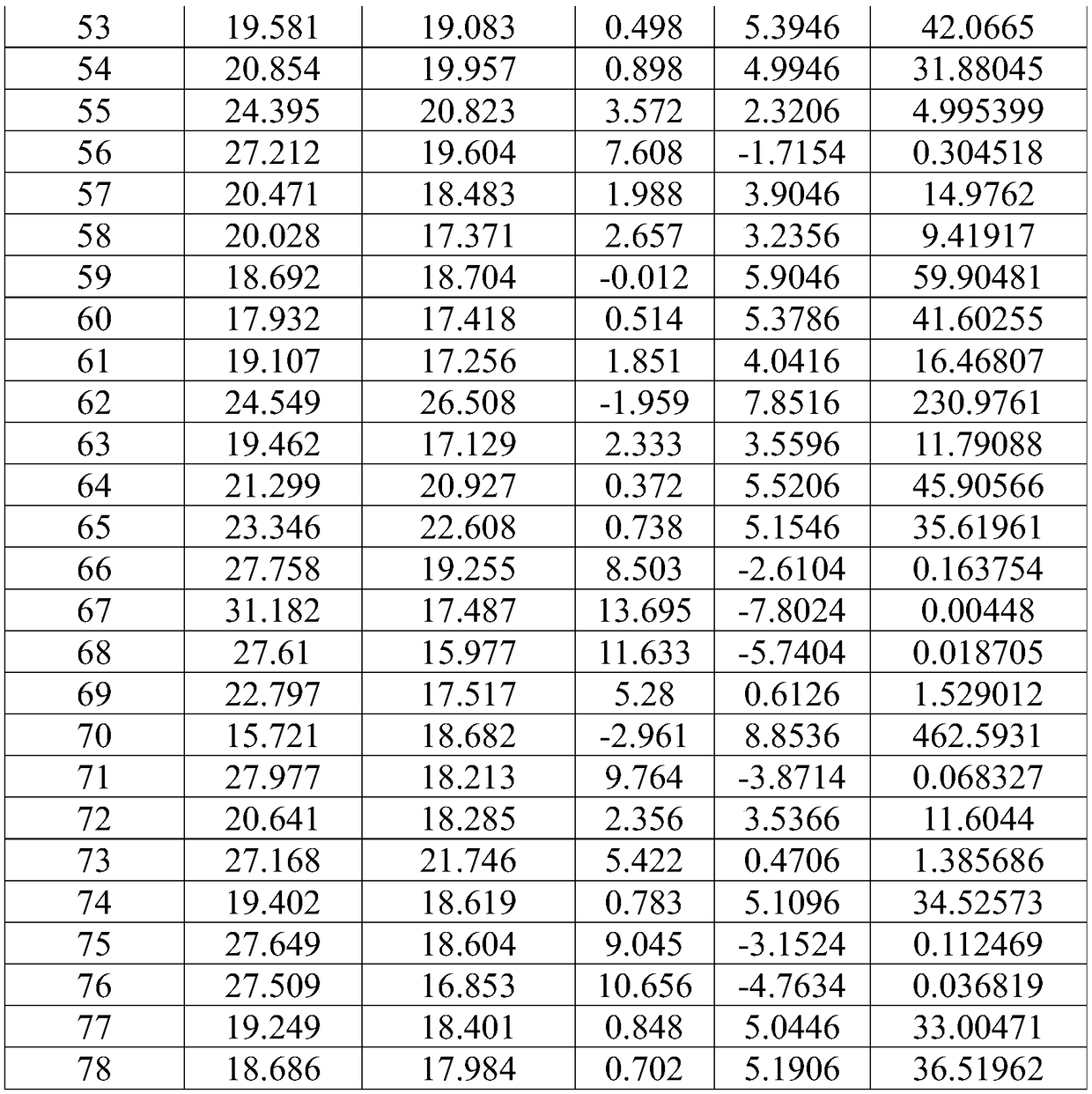
[0108] A method for detecting CLC gene expression in nasal cavity exfoliated cells, comprising the following steps:
[0109] Step 1: Extract RNA from nasal exfoliated cells:
[0110] Step 1: Take the genomic DNA adsorption column and place it in a 2mL collection tube, dissolve the nasal exfoliated cells in 100 μL of cell lysate, add them to the genomic DNA adsorption column, centrifuge at 12,000 rpm for 60 seconds, and take the filtrate. Add an equal volume of 70% ethanol to the filter, mix well and add to the RNA purification column, centrifuge at 12000 rpm for 1 min, remove the filtrate,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com