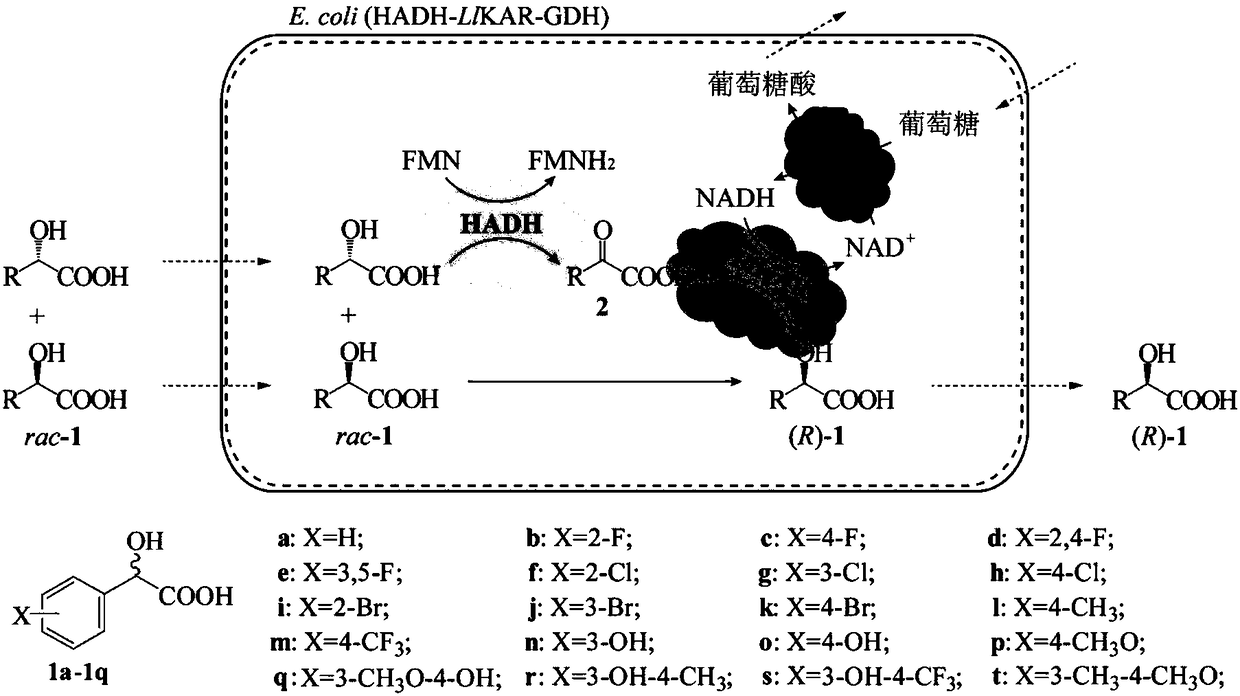
Keto acid reductase, gene, engineering bacterium and application of keto acid reductase in synthesis of chiral aromatic 2-hydroxy acid
A technology of reductase and engineering bacteria, applied in the direction of genetic engineering, oxidoreductase, application, etc., can solve the problems of limited catalytic efficiency, substrate loading and low yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Embodiment 1: screening efficient ketoacid reductase LlKAR
[0048]Using the amino acid sequence of the known ketoacid reductase LeKAR (shown in SEQ ID NO.2, the corresponding nucleotide sequence is shown in SEQID NO.1) as a template, five potential ketoacid reductions were obtained by NCBI-Blastp online alignment Enzyme sequences are respectively LlKAR (shown in amino acid sequence SEQ ID NO.4, nucleotide sequence shown in SEQ ID NO.3), LpKAR (shown in amino acid sequence SEQ ID NO.6, nucleotide sequence shown in SEQ ID shown in NO.5), LmKAR (shown in amino acid sequence SEQ ID NO.8, nucleotide sequence shown in SEQ ID NO.7), KoKAR (shown in amino acid sequence SEQ ID NO.10, nucleotide sequence is Shown in SEQ ID NO.9) and SnKAR (shown in amino acid sequence SEQ ID NO.12, nucleotide sequence shown in SEQ ID NO.11), the amino acid sequence homology is respectively 84%, 78%, 74%, 49% and 49%. Subsequently, the corresponding nucleotide sequences were synthesized in vitr...
Embodiment 2
[0055] Embodiment 2: Construction of recombinant bacteria E.coli BL21(DE3) / pET28b-HADH / pCDFDuet-LlKAR-GDH
[0056] Glucose dehydrogenase (GDH) gene (nucleotide sequence shown in SEQ ID NO.15, amino acid sequence shown in SEQ ID NO.16) derived from Exiguobacterium sibiricum (WP_012369122.1) was synthesized corresponding nucleosides in vitro The acid sequence was inserted into the expression plasmid pET28b to obtain the recombinant plasmid pET28b-GDH, which was transformed into E.coli BL21(DE3), spread on the LB plate containing 50 μg / mL kanamycin, and screened the recombinant bacteria E. coli BL21(DE3) / pET28b-GDH.
[0057] With the seamless cloning kit ( II, Vazyme Biotech Co., Ltd), the nucleotide sequence of GDH and the preferred L1KAR were connected to the expression vector pCDFDuet-1 successively to obtain the recombinant plasmid pCDFDuet-L1KAR-GDH, which was transformed into E.coli BL21(DE3), Spread on LB plates containing 50 μg / mL streptomycin to screen recombinant ba...
Embodiment 3
[0061] Example 3: Inducing the co-expression of recombinant bacteria E.coli BL21(DE3) / pCDFDuet-LlKAR-GDH dual enzymes
[0062] Inoculate recombinant Escherichia coli E. coli BL21(DE3) / pCDFDuet-LlKAR-GDH into LB liquid medium containing 50 μg / mL streptomycin, shake and culture at 37°C and 150 rpm for 8-10 hours to obtain seed liquid; Put the seed solution into the LB liquid medium containing 50 μg / mL streptomycin according to the inoculum amount of 2% (volume concentration), and cultivate it with shaking at 37°C and 150rpm until the OD 600When it reaches 0.6, add IPTG to a final concentration of 0.1mM, shake and culture at 28°C and 150rpm for 10-12h, collect the wet cells by centrifugation and wash twice with saline to obtain E.coli BL21(DE3) / pCDFDuet-LlKAR - Resting cells of GDH. Using uninduced E.coli BL21(DE3) / pCDFDuet-LlKAR-GDH as a control, SDS-PAGE protein electrophoresis analysis was carried out, and the results were as follows Figure 4 As shown, it can be seen that a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com