Enzymatic hydrolysis peptide map analysis method for polyethylene glycol modified protein medicine
A technology of polyethylene glycol and analysis method, which is applied in the field of peptide map analysis, and can solve problems such as the absence of PEG-modified peptides and incomplete enzymatic hydrolysis.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
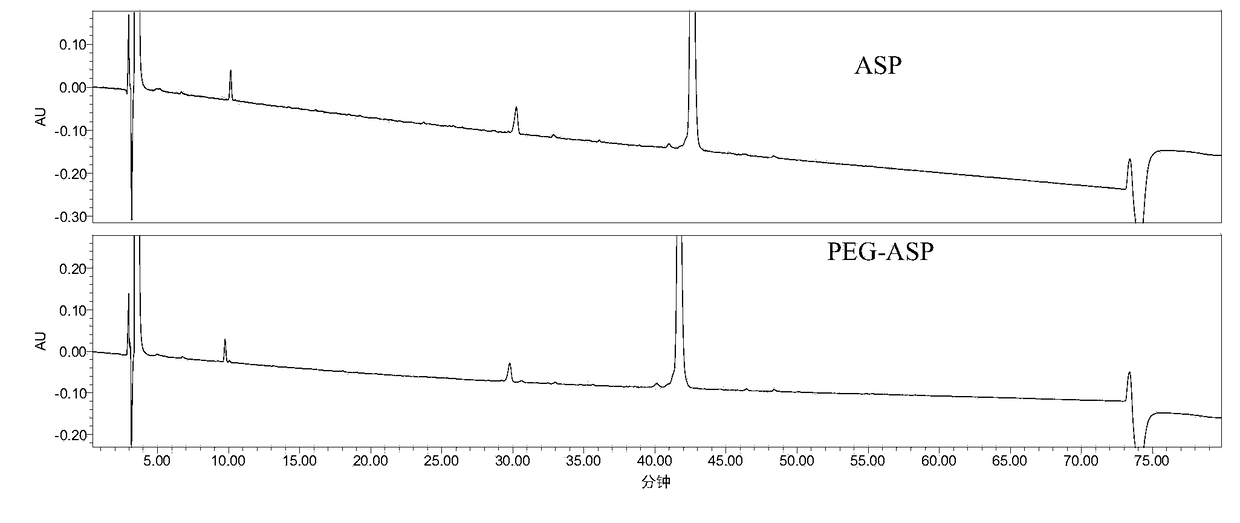
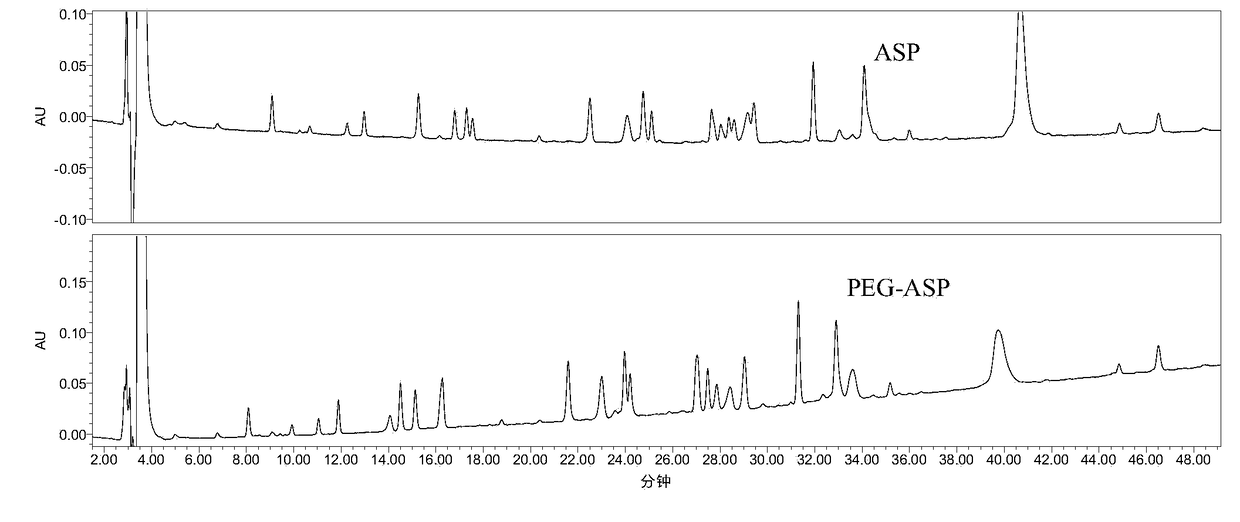
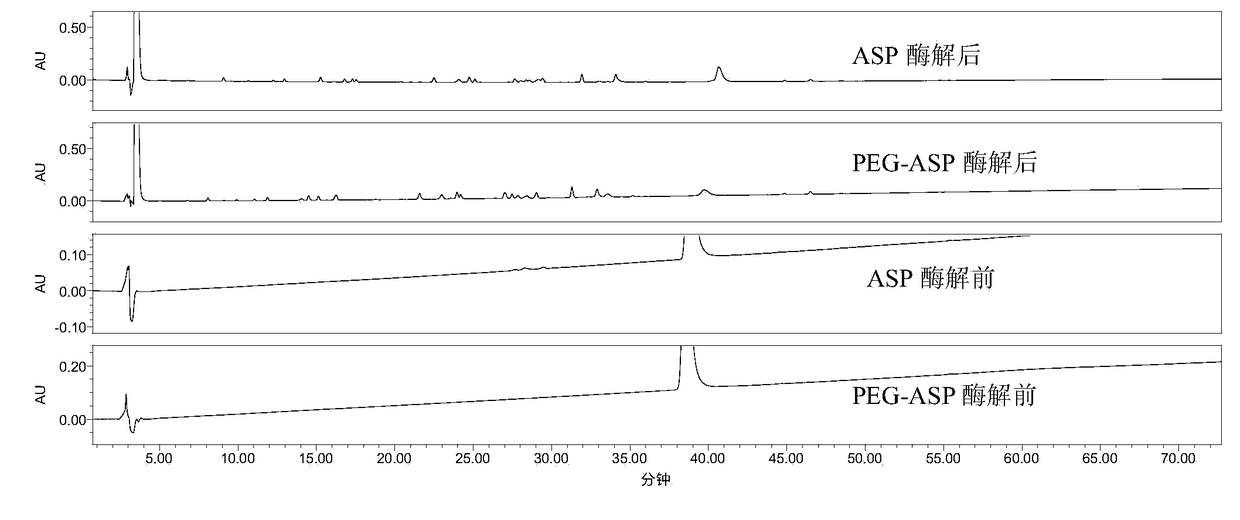
[0078] Example 1 The peptide map analysis method of the present invention is applied to ASP and PEG-ASP
[0079] 1. Detection method
[0080] (1), enzymolysis:
[0081] The sample to be tested is desalted and replaced with 1% NH 4 HCO 3 In buffer (PH7.8);
[0082] Dilute the sample to 1.0 mg / mL, take 100 μL, and heat at 100 °C for 5 min;
[0083] 2ug trypsin (1mg / ml, add 2μl) per 100ug heated sample solution, digest at 37℃ for 16-24h;
[0084] Termination of the reaction: 10 μl of 50% acetic acid solution was added to the reaction system at 1:10 (v / v) to terminate the reaction;
[0085] PEG-ASP enzymatic hydrolysis blank and ASP enzymatic hydrolysis blank (that is, unenzymatically hydrolyzed PEG-ASP and ASP samples were prepared in the same way, with 2 μL of 1% NH 4 HCO 3 buffer (pH ~7.8) in place of 2 μL of enzyme).
[0086] (2), RP-HPLC detection
[0087] Chromatographic column: XBridge Protein BEH C4 column (4.6×250mm,
[0088] Mobile phase: A: 0.1% TFA + water / ...
Embodiment 2
[0100] Example 2 The peptide map analysis method of the present invention is applied to polyethylene glycol modified arginine deiminase (PEG-ADI)
[0101]1. Enzymatic hydrolysis:
[0102] To 500 μL of PEG-ADI sample, 20 μL of 0.5mM DTT and 20 μL of 2M urea were added, and the reaction was denatured at room temperature for 30 min. Desalination replacement with 1% NH 4 HCO 3 In the solution, 100 μL was taken and heated at 100 °C for 5 min. After cooling, 2 μg trypsin was added, and the enzyme was hydrolyzed at 37°C for 16 to 24 hours. The reaction was terminated by adding 10 μL of 50% acetic acid to the system at 1:10 (V / V). The processed samples were centrifuged for later use.
[0103] 2. RP-HPLC detection:
[0104] Chromatographic conditions:
[0105] Mobile phase: A: 0.1% TFA + water / acetonitrile (95:5, v / v); B: 0.1% TFA + water / acetonitrile (35:65, v / v)
[0106] Chromatographic column: XBridge Protein BEH C4 column (4.6×250mm,
[0107] The gradients are shown in T...
Embodiment 3
[0115] Example 3 The peptide map analysis method of the present invention is applied to polyethylene glycol-modified KLK1 (PEG-KLK1)
[0116] 1. Enzymatic hydrolysis
[0117] 1) Concentrate the enzyme solution containing the PEG-KLK1 sample by centrifugal ultrafiltration, centrifuge at 4000 rpm for 30 min, take 0.3 M Tris-HCl (pH 7.8, containing 1 M TCEP HCl and 10 mM EDTA) solution to resuspend the protein, dissolve, and resuspend the protein. The sample concentration was brought to about 1.0 mg / mL.
[0118] 2) Add 1:25 (v / v) μL of 0.5M iodoacetic acid to the reaction system, and react at room temperature for 30 minutes in the dark.
[0119] 3) Add 1:50 (v / v) μL of 0.5M DTT to the reaction system and mix well.
[0120] 4) The above mixture was taken and passed through a G25Desalting column for desalting, and replaced in Tris-HCl (pH 7.8) buffer solution containing 2M urea.
[0121] 5) Take 100 μL of the desalted mixture, add 2 μg trypsin at a ratio of 1:50 (v / v) (dissolve ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com