Method for quantitatively assaying serum miRNA by utilizing RNase ONE nuclease and chemiluminescence technology
A technology of chemiluminescence and ribonuclease, applied in chemiluminescence/bioluminescence, analysis through chemical reaction of materials, preparation of test samples, etc., can solve problems such as inability to meet clinical detection requirements and complicated operation, and achieve Meet clinical detection needs, high sensitivity, and prolong the effect of detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1~8
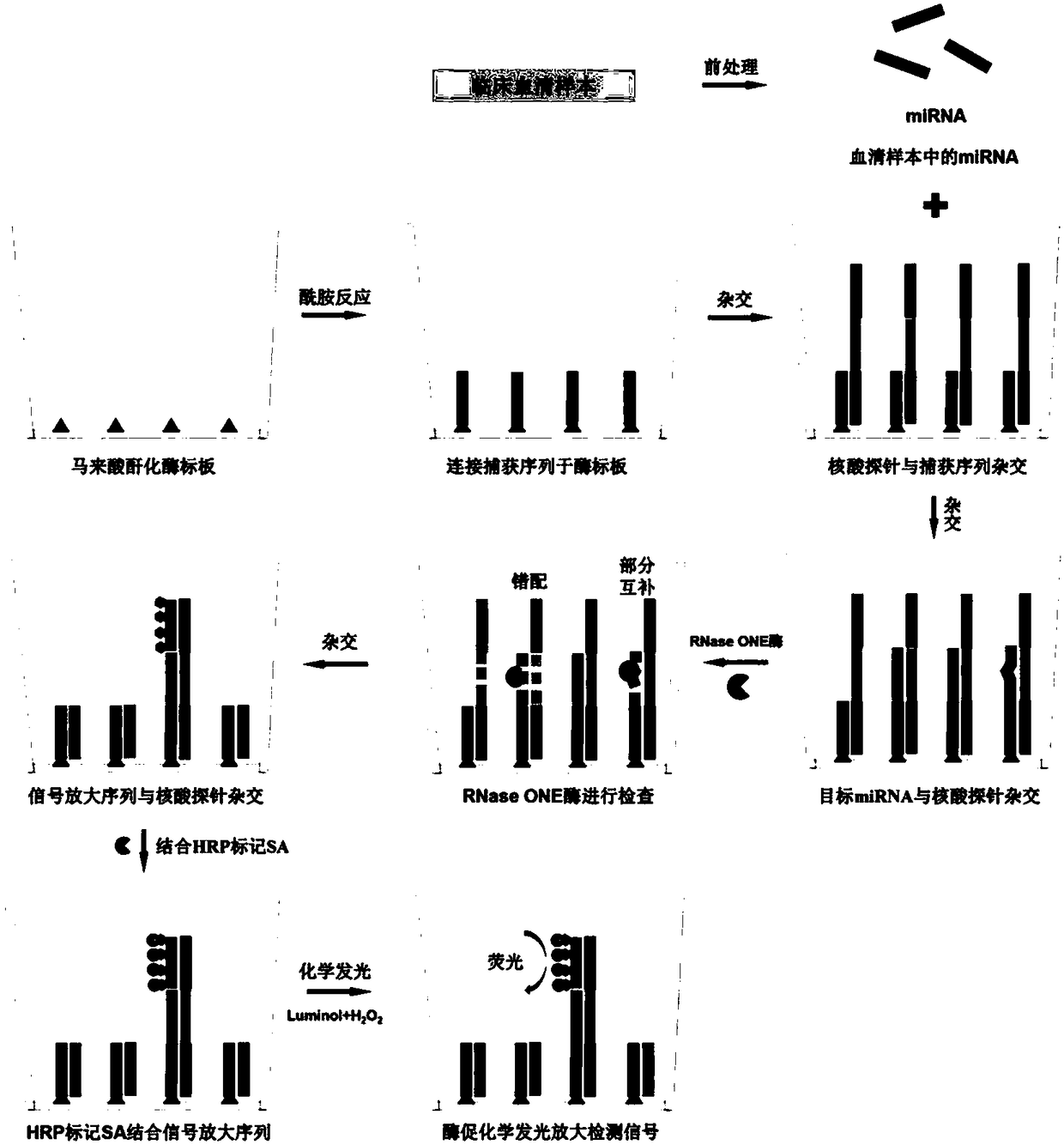
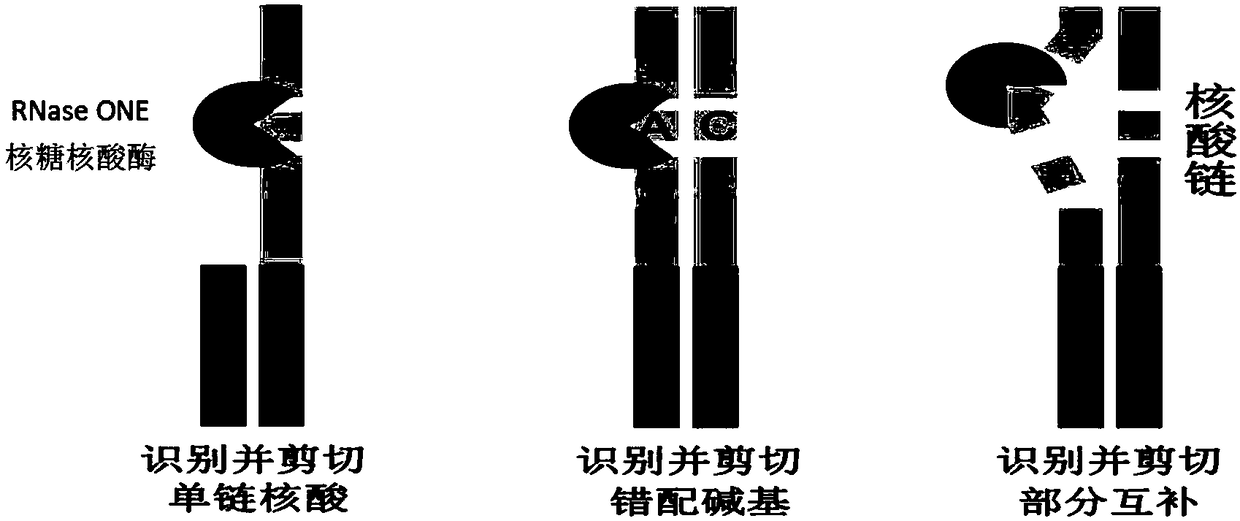
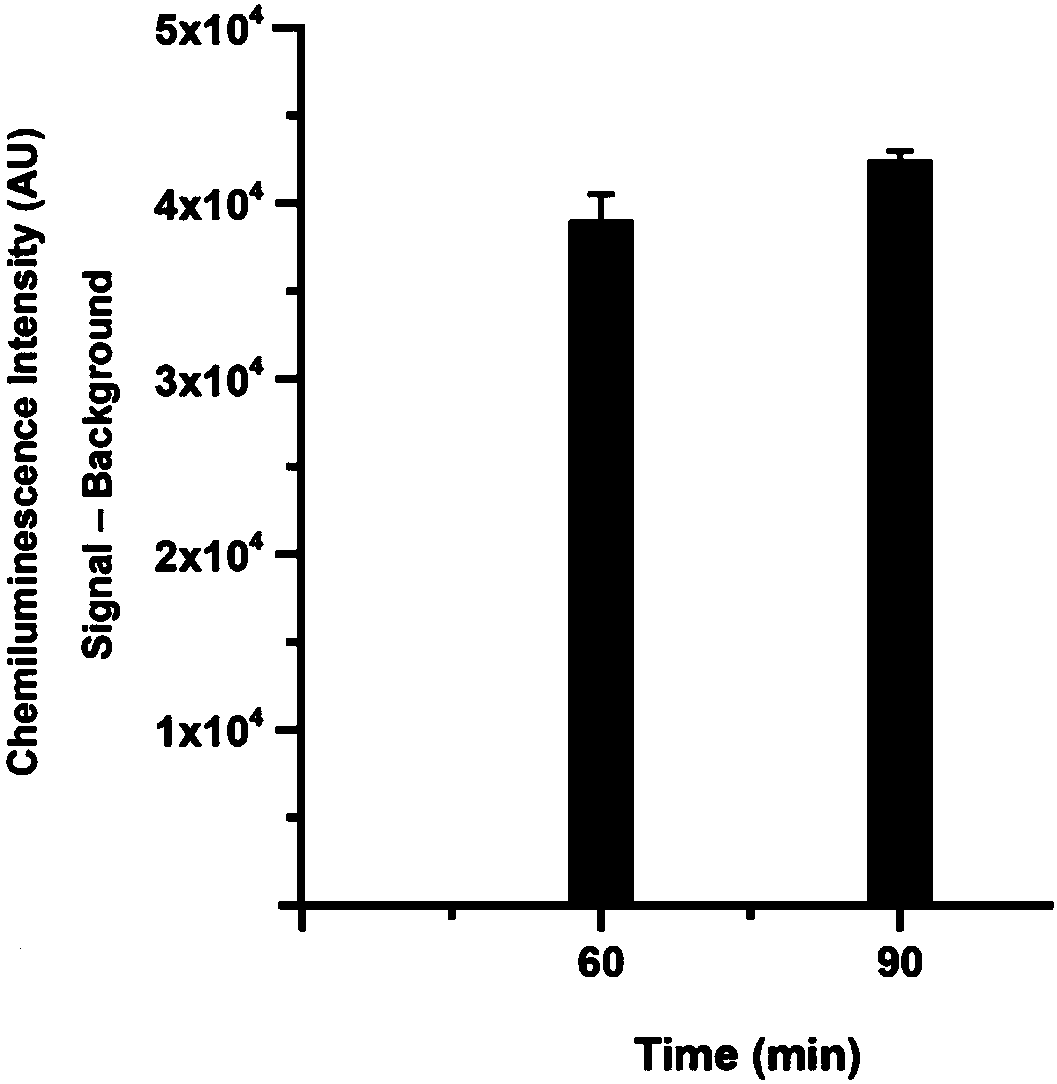
[0053] Examples 1-8 provide a method for quantitative detection of serum miRNA markers using RNase ONE ribonuclease and chemiluminescence technology. Such as figure 1 As shown, the method is as follows:
[0054] (1) Pretreatment of serum samples
[0055] The Tween 20 (10%) solution was diluted with TE buffer (pH 8.0) to a final concentration of 1%, and then Proteinase K was added to a final concentration of 200 μg / ml to prepare serum treatment solution. After the serum sample is thawed, vortex 5 to 10 times. The prepared treatment solution and serum samples were respectively placed in a constant temperature mixer at 37° C. for 30 minutes with slight shaking and mixing. The serum sample and the treatment solution were mixed at a volume ratio of 1:1, and reacted at 60° C. at a speed of 300 rpm for 60 to 90 minutes. After processing, the serum solution should be used for the next reaction immediately, or stored in a -80°C refrigerator.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com