Synthetic method of lipopeptide based on pseudomonas chlororaphis
A technology of Pseudomonas chloropinus and synthetic methods, applied in the direction of microorganism-based methods, biochemical equipment and methods, microorganisms, etc., can solve problems that are not clearly given
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
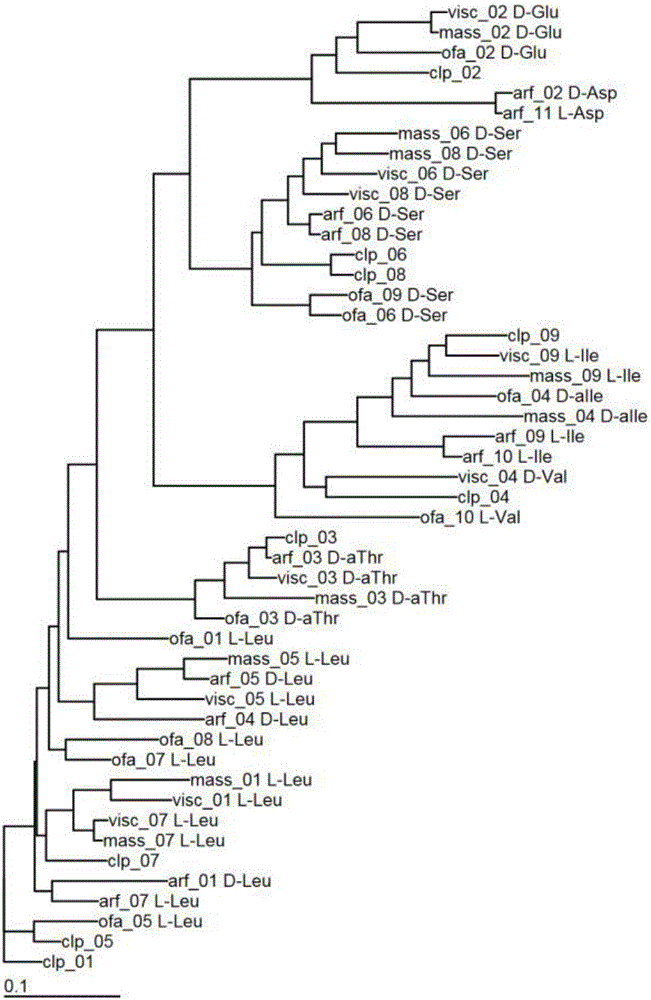
[0033] Example 1: Cluster analysis of lipopeptide synthetic proteins
[0034]Select the lipopeptide synthesis protein sequence of Pseudomonas chloropinus HT66 and the lipopeptide synthesis protein sequence of known structure in other Pseudomonas, conduct module analysis, and construct a phylogenetic tree; according to the homology and The position determines the corresponding amino acid and its sequence. For protein sequence determination, refer to literature (Gross H, Stockwell VO, Henkels MD, et al. The genomisotopic approach: a systematic method to isolate products of orphan biosynthetic gene clusters [J]. Chemistry & Biology, 2007, 14(1): 53-63) .
[0035] Protein sequence analysis found that the lipopeptide synthesis protein of strain HT66 contained 9 modules, indicating that the lipopeptide product was composed of 9 amino acids. From the NCBI database, the inventor found 4 strains of Pseudomonas that can synthesize lipopeptides and whose molecular structure has been ...
Embodiment 2
[0039] Example 2: Construction of lipopeptide synthesis gene cluster deletion strain
[0040] Primers were designed based on the whole genome sequence of strain HT66. The primer pair (5'-3') for amplifying the upstream homology arm of the lipopeptide synthesis gene cluster clp is:
[0041] clp-F1 (SEQ ID NO. 1): CG GAATTC AGCATCTTCGCCGCAAACAG (EcoRI),
[0042] clp-R1 (SEQ ID NO. 2): CCGGGATGAATGGGTAGTCG;
[0043] The primer pair for amplifying the downstream homology arm of clp is:
[0044] clp-F2 (SEQ ID NO. 3): ACTACCCATTCATCCCGGCGGGCTATGGATGGGTTCG,
[0045] clp-R2 (SEQ ID NO. 4): C GGATCC TGGAGATCGCCGCGTCTTC (BamHI).
[0046] The PCR system and procedures were carried out with reference to the literature (Gross H, Stockwell VO, Henkels MD, et al.Thegenomisotopic approach: a systematic method to isolate products of orphanbiosynthetic gene clusters[J].Chemistry&Biology,2007,14(1):53-63) . Genomic DNA extraction, plasmid extraction, electrophoresis gel fragment rec...
Embodiment 3
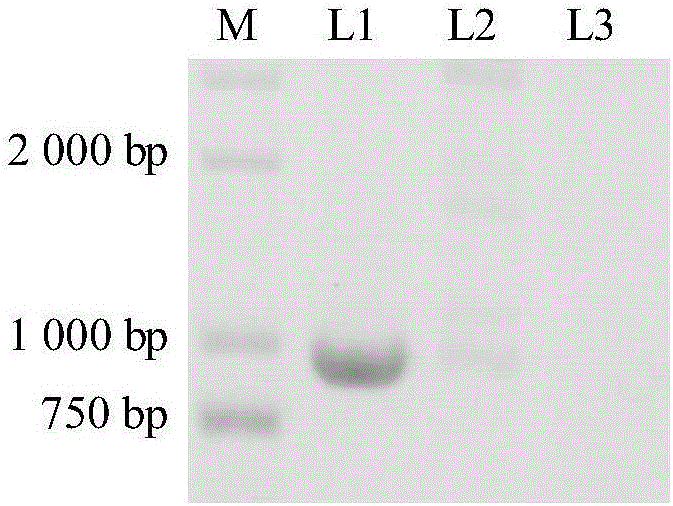
[0048] Embodiment 3: the mass spectrometry analysis of lipopeptide product
[0049] The strains HT66 and HT66Δclp were fermented in KB medium respectively, and the lipopeptide products were extracted from the 24h fermentation broth using chloroform-methanol (2:1) solution, and the organic phase was taken and dried with nitrogen (Smyth T, Rudden M, Tsaousi K, et al. Protocols for the isolation and analysis of lipopeptides and bioemulsifiers [M]. New York; Humana Press. 2014:1-26). The crude extract was dissolved in acetonitrile, filtered and detected by ultra-high performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC / QTOF-MS). The instrument is WatersACQUITYTM UPLC&Q-TOF MS Premier. The chromatographic column is Waters CORTECS 1.6μm (2.1mm×100mm), the column temperature is 45°C, the injection volume is 1μL, the flow rate is 0.4mL / min, the detection wavelength is 254nm and 210nm; the gradient elution conditions are: 0min, water 95 %, acetonitril...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com