Method of increasing yield of lignocellulose substrate hydrolase
A lignocellulose and cellulase technology, applied in the field of bioengineering, can solve the problems of reduced specific enzyme activity, large enzyme dosage, low enzymatic hydrolysis efficiency of cellulase system, etc., and achieves wide application value, increased copy number, high efficiency expressive effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
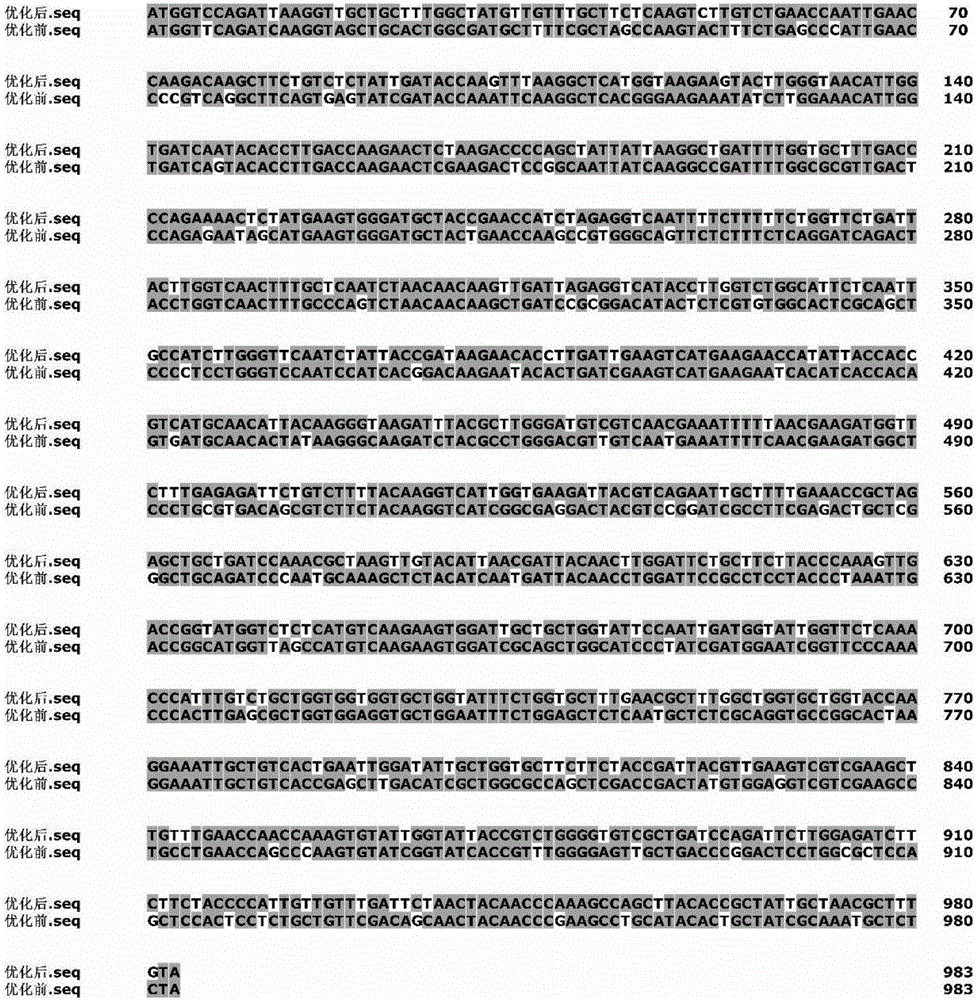
[0033] Example 1: Codon optimization of xylanase (XynC) gene
[0034] Search the Aspergillus niger xylanase gene sequence (accession number: KJ601783) in GenBank of NCBI (http: / / www.ncbi.nlm.nih.gov / ), and perform codon optimization according to the following method:
[0035] (1) Using the software GeneDesigner 2.0, the XynC codon (SEQ ID NO.1) was initially replaced according to Pichia pastoris codon preference; and the GC content was calculated, and then further adjusted according to the GC content to make the value within 40 ~50%. Analyze the distribution of hidden splice sites and codons of mRNA in the adjusted gene sequence. With the help of this software, the amount of change before and after codons can be visually analyzed.
[0036] (2) Using RNAStructure 3.2 to predict and analyze the free energy of the start codon end of the mRNA secondary structure, select a sequence with lower free energy and an open-loop structure at the start codon end, and finally obtain the optimized ...
Embodiment 2
[0039] Example 2: Construction of recombinant expression vectors pPIC9K-AxynC and pPICZαA-AxynC
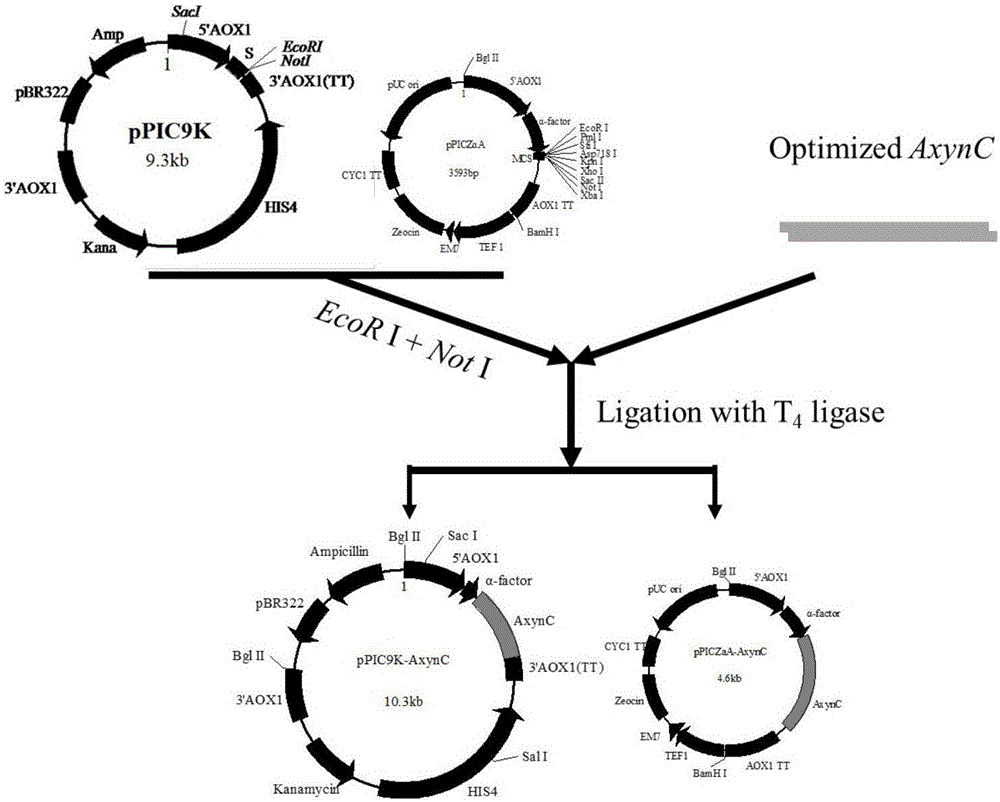
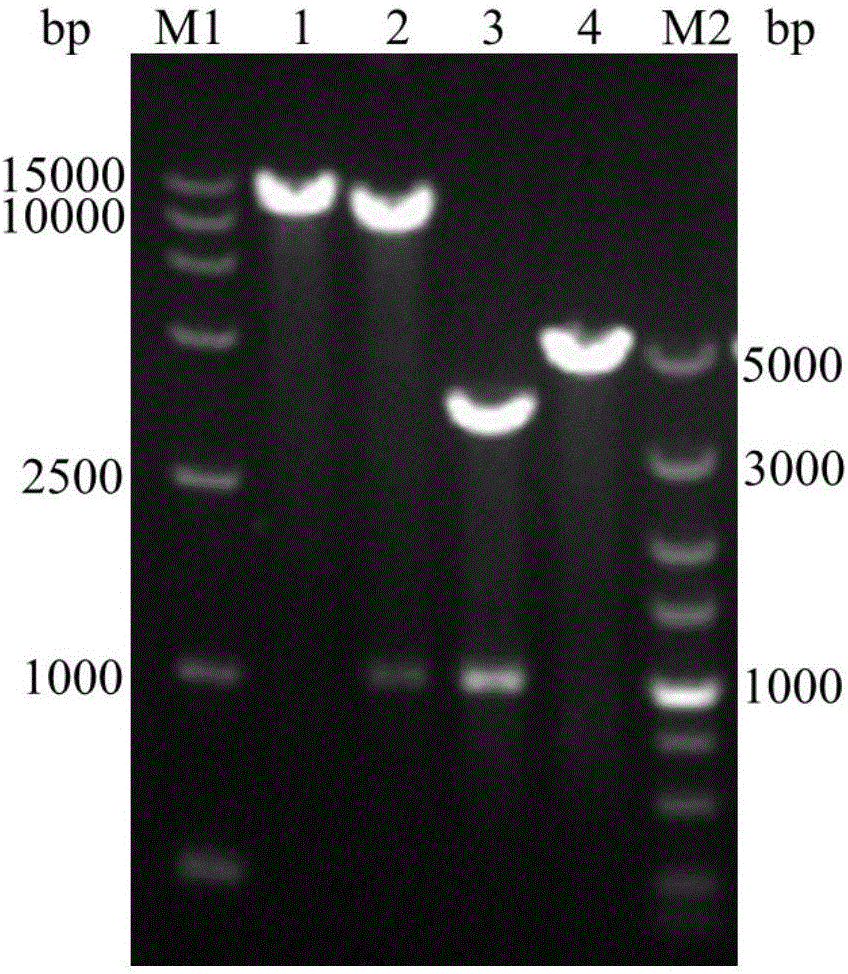
[0040] The expression vectors pPIC9K and pPICZαA were digested with EcoR I and Not I, and the digested products were recovered. After purification, the synthesized target gene and the vector pPIC9K and pPICZαA use T 4 DNA ligase was used for ligation and transformed into Escherichia coli DH5α, and the target vectors pPIC9K-AxynC and pPICZαA-AxynC were obtained by colony PCR identification. Cultivate the Escherichia coli containing the target vector in LB medium for 12 to 16 hours, and extract the target vector according to the plasmid extraction manual. The construction and verification of expression vectors pPIC9K-AxynC and pPICZαA-AxynC are as follows figure 2 with image 3 Shown.
Embodiment 3
[0041] Example 3: Construction of a recombinant Pichia pastoris with high xylanase production and strain screening
[0042] Sac I was used to linearize pPIC9K-AxynC, and electroporation was used to transform Pichia pastoris GS115 competent cells. The transformation liquid was coated with RDB solid medium and cultured at 30°C for 3 days. Pick the colonies that grow normally and transfer them to YPD plates containing different concentrations (1, 2, 4, and 6 mg / mL) of geneticin G418, and pick the colonies that can grow normally on the high-concentration plates. The colonies obtained above were respectively inoculated into 250mL Erlenmeyer flasks of 30mL BMMY medium, cultured at 30°C and 220rpm for 4 days, and methanol was added every 24h to make the final concentration 0.5%. After the culture, the supernatant was collected by centrifugation. The enzyme activity was measured with beech xylan as the substrate, and the bacteria with the highest enzyme activity was No. 16 (named P.past...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com