Production method for tagatose
一种生产方法、塔格糖的技术,应用在差向异构生产塔格糖领域,能够解决未能公开塔格糖等问题,达到降低生产成本、高产率的效果
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
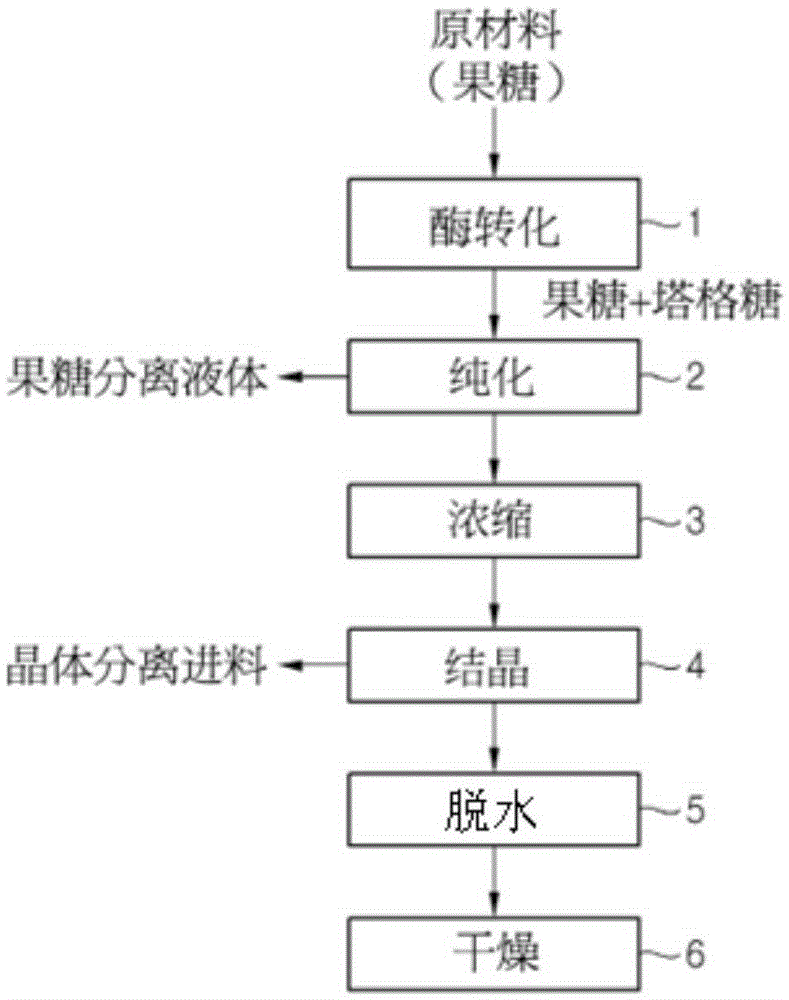
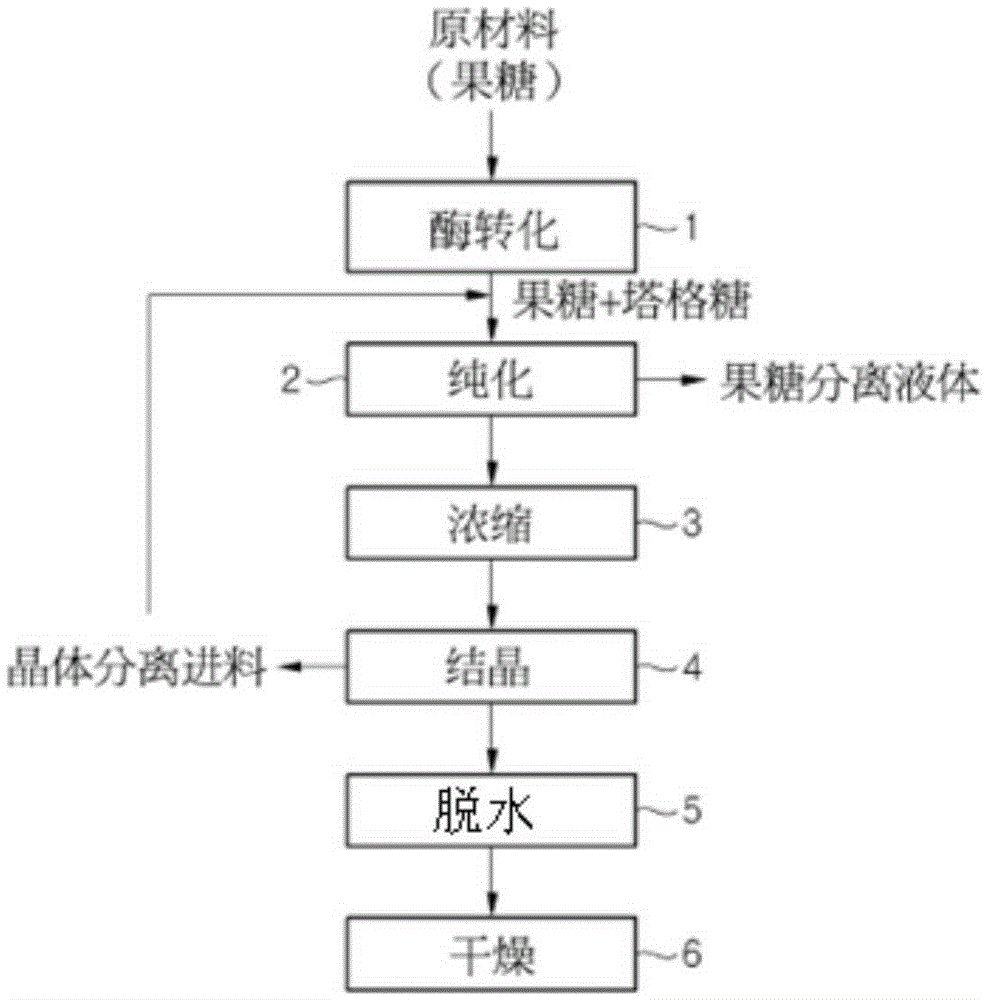
[0060] Evaluation of Chromatographic Separation Characteristics Based on Enzyme Conversion
[0061] In order to separate and crystallize a target material (tagatose) with high purity from mixed sugars (fructose and tagatose), purification using chromatographic separation is extremely important. In general, different recovery yields (%) are achieved for purified sugar solutions to obtain crystallized products, depending on the content of the target material (tagatose). High recovery yields (%) are obtained proportionally with increasing content of target material (tagatose). Thus, it is possible to produce the final product with relatively low production and processing costs. In order to efficiently produce crystalline sugar (high purity, high yield), it is extremely important to ensure that the purity of the target material (tagatose) contained in the feed (Feed) is 90% or more, more preferably 95%.
[0062] The operating conditions of chromatography can produce various resu...
example 2
[0074] Preparation of Ketohexose C4-epimerase
[0075] Genomic DNA (SEQ ID NO. 1) of Thermotoga maritima (strain ATCC43589 / MSB8 / DSM3109 / JCM10099) was used as a template, a thermotolerant microorganism and forward primers (5'-GGGCATATGATGGTCTTGAAAGTGTTCAAAG-3') and reverse Primer (5'-AAACTCGAGCCCCTCCAGCAGATCCACGTG-3') was used for PCR.
[0076] In addition, by using genomic DNA (SEQ ID NO: 2) of Thermotoga neoapollinii (DSM4359) as a template, a heat-resistant microorganism and adding a forward primer (5'-GGGCATATGATGGTCTTGAAAGTGTTCAAAG-3') and a reverse primer ( 5'-AAACTCGAGTCACCCCCTTCAACAGGTCTACGTG-3') for PCR.
[0077] Conditions of PCR are shown in Table 3 and Table 4. The gene amplified by PCR was inserted into pET-21a vector, followed by transformation into E. coli DH5α strain. Plasmids were isolated from transformed strains, which were sequenced to identify the base sequence of the inserted gene. The plasmid was transformed into the protein expression strain Escheric...
example 3
[0085] Purification of Ketohexose C4-epimerase
[0086] In order to measure the activity of ketohexose C4-epimerase expressed in microorganisms, protein purification was performed by the following method. The culture solution that completed the protein expression was centrifuged at 8,000rpm for 10 minutes to collect the microorganisms, and the microorganisms were resuspended in 50mM NaH containing 10mM imidazole (imidazole) and 300mMNaCl 2 PO 4 (pH8.0) buffer. Suspended microorganisms were disrupted by a sonicator and centrifuged at 13,000 rpm for 10 minutes to collect a supernatant. The collected supernatant was passed through a chromatography column packed with Ni-NTA resin as a filler. At this point, 50 mM NaH containing 20 mM imidazole and 300 mM NaCl 2 PO 4 (pH8.0) buffer flowed through in an amount of 10 times the volume of the filler in the chromatographic column, thereby removing proteins non-specifically attached to the filler. Finally, 50 mM NaH containing 250 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com