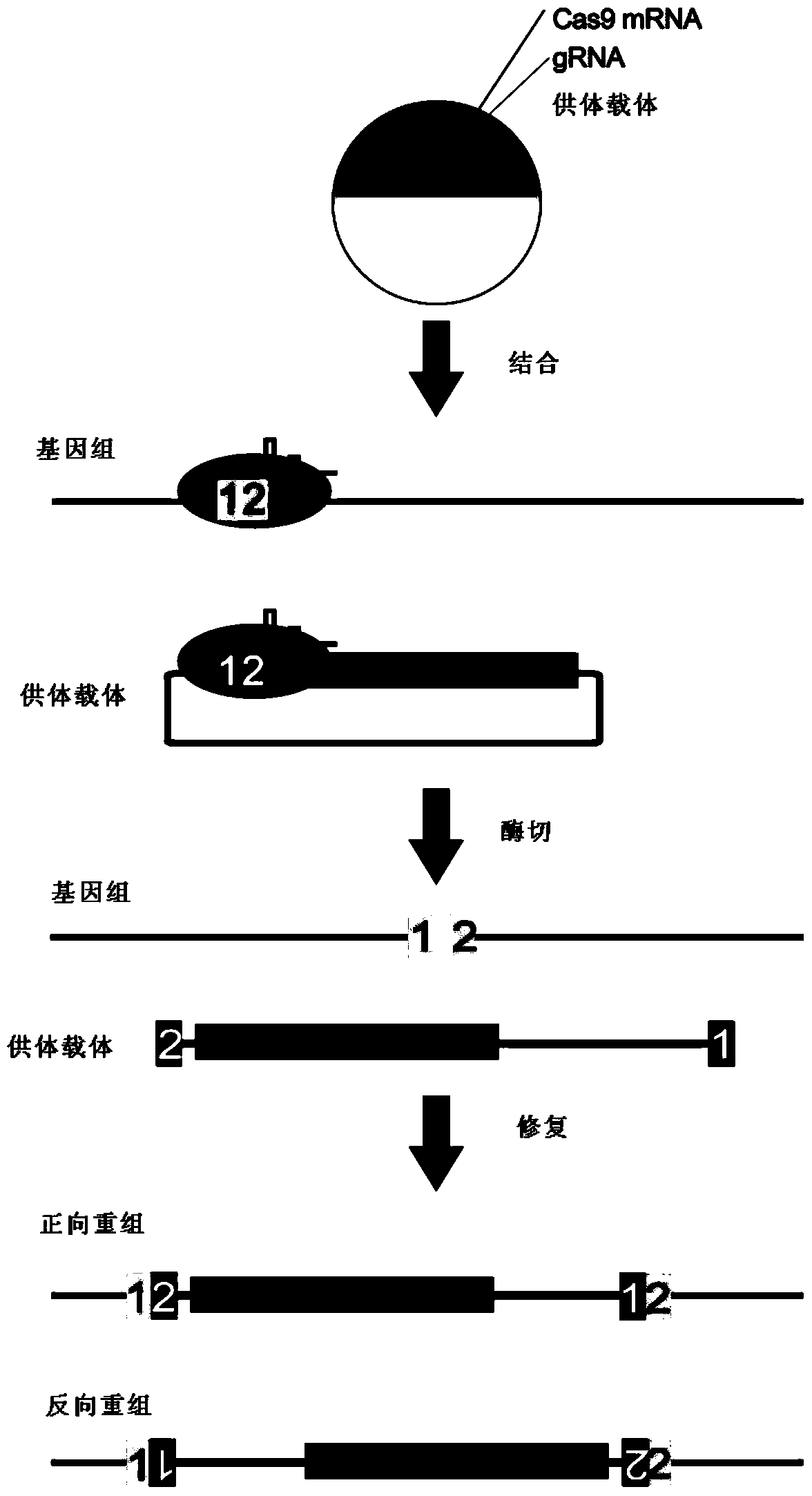
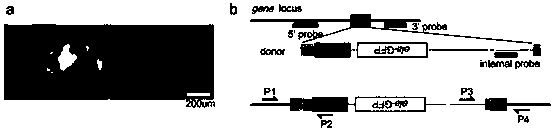Vector without frameshift mutation after recombination, method and application of site-directed gene knock-in in Xenopus frog genome
A genome and clawed frog technology, applied in the field of genetic engineering, can solve the problems of cumbersome, low efficiency, and inability to do long-term genetic research.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0093] Example 1 Gene site-specific insertion in exon 5 of the ets1 gene in Xenopus tropicalis
[0094] 1. Cas9 target site
[0095] The ets1 target is the target site corresponding to ets1-T2 in Table 1 in the literature (Guo et al., 2014) (the sequence is shown in SEQUENCE NO.15, that is, 5-GGTTCAGAGAATTCAGAGGGCGG-3)
[0096] 2. Preparation of Cas9mRNA and gRNA
[0097] Synthesize Cas9 mRNA and gRNA according to the following scheme:
[0098] 1) Preparation of Cas9 mRNA
[0099] Endonuclease NotI will 10ug pCS2-3×FLAG-NLS-SpCas9-NLS (addgene ID: 51307; vector structure as image 3 Shown in a) The plasmid was linearized, and Cas9 mRNA was prepared by in vitro transcription using SP6.
[0100] 2) gRNA preparation
[0101] a. Vector preparation
[0102] Referring to the method of the literature (Guo et al., 2014), the Cas9 target site shown in SEQUENCE NO.15 was cloned into the backbone vector by using the enzyme digestion effect of BbsI, and the backbone vector was pUC57...
example 2
[0131] Example 2 Carrying out site-directed gene insertion in exon 3 of the ets2 gene in Xenopus tropicalis
[0132] 1. Cas9 target site
[0133] The ets2 target is the target site corresponding to ets2 in Table 1 in the literature (Guo et al., 2014) (the sequence is shown in SEQUENCE NO.25), that is, ggtctggact cttactctca tgg.
[0134] 2. Preparation of Cas9mRNA and gRNA
[0135] 1. Preparation of Cas9 mRNA, see Example 1
[0136] 2. Preparation of gRNA, see Example 1
[0137] a. Vector preparation
[0138] Referring to the method of the literature (Guo et al., 2014), the Cas9 target site as shown in SEQUENCE NO.25 was cloned into the backbone vector by using the enzyme digestion effect of BbsI, and the backbone vector was pUC57-T7-gRNA plasmid (vector structured as image 3 b) to obtain ets2-specific gRNA.
[0139] b. gRNA transcription synthesis, see Example 1
[0140] 3.donor preparation
[0141] 1) PCR amplification
[0142] The genome sequence (611bp) containing...
example 3
[0157] Example 3 Carrying out gene-directed insertion at intron 1 of the tropical clawed frog tyrosinase gene to achieve the effect of no frame shift
[0158] 1. Preparation of Cas9 mRNA and gRNA
[0159] 1) Cas9mRNA synthesis method is the same as in Example 1
[0160] 2) Preparation of gRNA
[0161] The target site is within intron 1, about 600bp away from exon 2, named tyr-int1.
[0162] The target site recognition sequence is 5-GGGGTCCCTAACTTCCTCTATGG-3 (SEQUENCE NO.31).
[0163] The two annealed single-stranded sequences are as follows:
[0164] tyr-int1-S: 5-TAGGGGTCCCTAACTTCCTCTA-3 (SEQUENCE NO.32)
[0165] tyr-int1-A: 5-AAACTAGAGGAAGTTAGGGACC-3 (SEQUENCE NO.33)
[0166] The gRNA synthesis method is the same as Example 1.
[0167] 2.donor preparation
[0168] 1) PCR amplification
[0169] Using PCR amplification (template is the genome of Xenopus tropicalis) to obtain the partial sequence of the intron containing the tyr-int1 target site and the exon adjacent to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com