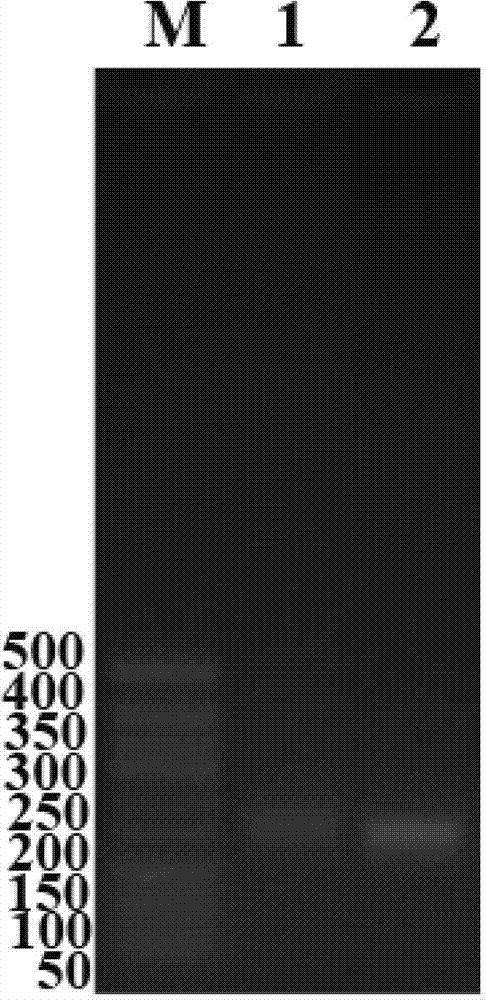
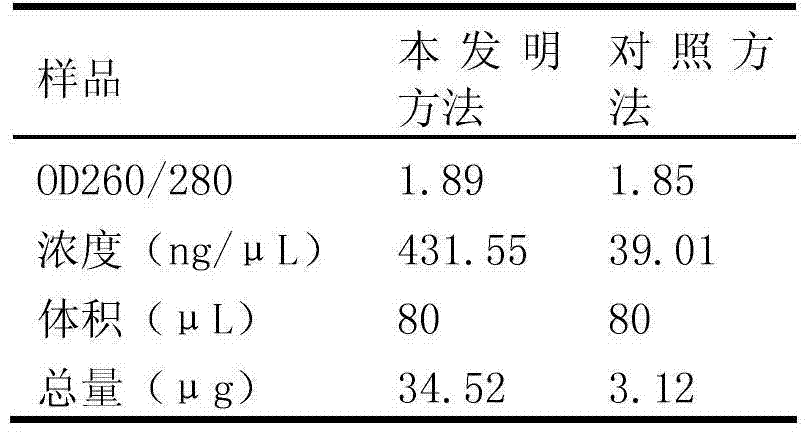
Method for extracting the total microbial genome in vinegar fermentation process
A fermentation process and microbial technology, applied in the field of molecular biology, can solve the problems that the quality and effect of extraction cannot be guaranteed, and achieve the effect of ensuring integrity and high quality of total DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] The sample in this embodiment is a sample of vinegar solid vinegar fermented grains.
[0027] (1) Take 2 g of vinegar unstrained spirits sample and crush it into fine particles in a sterile mortar, add liquid nitrogen and quickly grind it to powder.
[0028] (2) Take 0.3g sample and add 2mL DNA extraction solution, place it on a constant temperature shaking shaker at 37°C and shake at 300rpm for 20min, then add 200μL 2% SDS, vortex and mix well, then quickly place it in a water bath at 65°C for 1h Gently invert several times every 15min-20min, add PEG8000 (20%-40%, dissolved in 1.6mol / L NaCl, volume ratio) to the extract solution and place at room temperature for 2h, centrifuge at 10000×g for 130min, discard the supernatant, Dissolve the precipitate in TE buffer (10mmol / L Tris-HCl, 1mmol / L EDTA, pH8.0), add 3mol / L potassium acetate solution, place at 4°C for 60min, and then centrifuge at 13000×g for 15min at room temperature.
[0029] (3) Take 1 mL of supernatant aqueo...
Embodiment 2
[0039] The sample in this case is a sample of vinegar semi-solid wine mash.
[0040] (1) Take 5g of wine mash sample and centrifuge at 8000×g for 20min at 4°C, discard the supernatant, place the sample at -80°C for 30min, then vacuum freeze-dry for 2h, put the dried sample evenly into a sterile mortar, pour into the liquid Nitrogen is rapidly triturated until the sample becomes a white powder.
[0041] (2) Quickly weigh 0.5g of sample, add 1.5mL of DNA extraction buffer, place on a constant temperature shaking shaker at 37°C at 400rpm for 15min, add 250μL of 2% SDS, vortex to mix, and quickly place at 65 Incubate in a water bath at ℃ for 1 hour, gently invert several times every 15min-20min, add 2 times the volume of PEG8000 (20%-40%, dissolved in 1.6mol / L NaCl, volume ratio) to the extract and place at room temperature for 1h, Centrifuge at 12000×g for 10min, discard the supernatant, dissolve the precipitate in 2mL TE buffer (10mmol / L Tris-HCl, 1mmol / L EDTA, pH8.0), add 10% ...
Embodiment 3
[0051] (1) Weigh 0.3 g of the pretreated and ground sample in Example 2, add 1 mL of DNA extraction buffer, place on a constant temperature shaking shaker at 37°C, shake at 200 rpm for 30 min, add 200 μL of 2% SDS, and vortex to mix After homogenization, quickly place it in a 65°C water bath and incubate for 2 hours, gently invert it every 15 minutes, and add an equal volume of PEG8000 (20%-40%, dissolved in 1.6mol / L NaCl, volume ratio) to the extract Place at room temperature for 1.5h, centrifuge at 10000-12000×g for 10-30min, discard the supernatant, dissolve the precipitate in 1mL TE buffer (10mmol / L Tris-HCl, 1mmol / L EDTA, pH8.0), add 20% (v / v) 3mol / L potassium acetate solution, placed at 4°C for 15 minutes, and then centrifuged at 15000×g for 10 minutes.
[0052] (2) Take 1 mL of supernatant aqueous phase, add 1 mL of phenol: chloroform: isoamyl alcohol mixture (volume ratio 25: 24: 1), invert gently a few times, and centrifuge at 15000 × g for 10 min.
[0053] (3) Aspi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com