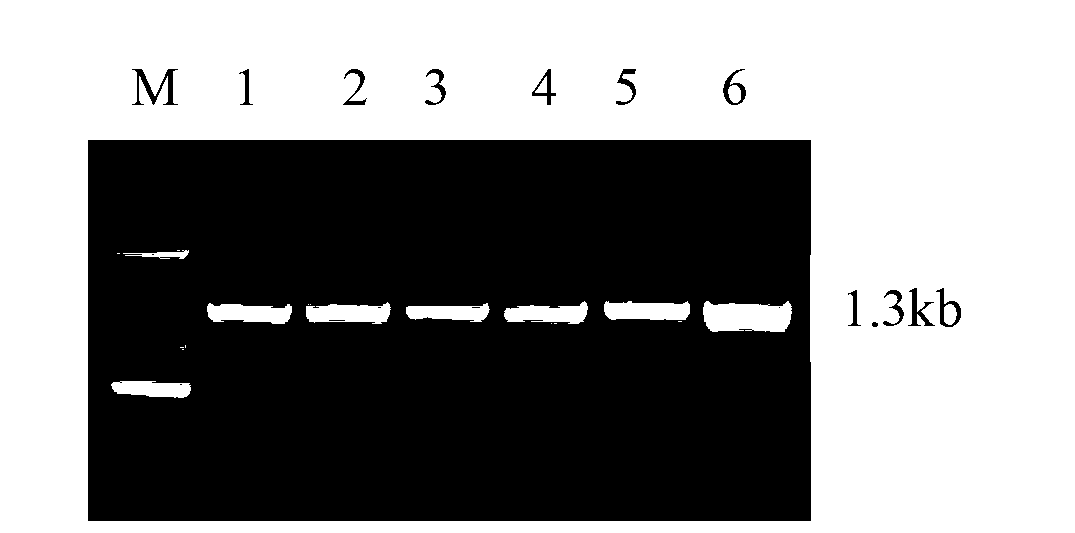Engineering bacteria for efficiently degrading two fungal toxins and application
A technology of mycotoxins and engineering bacteria, which is applied in the direction of fungi, bacteria, and microorganism-based methods, and can solve problems such as secondary pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1: Cloning of mycotoxin DON degrading enzyme gene DONase
[0032] Construction of Plasmid Library of Deoxynivalenol Degrading Enolase Gene
[0033] (1) The standard strain Fusarium oxysporum (Fusarium oxysporum) ACCC No.36245 (purchased from China Agricultural Microorganism Culture Collection Management Center) was activated and inserted into PDA liquid medium (potato 200g / L, glucose 20g / L, agar 20g / L, PH natural), and then the suspension of the strain was added to 100ml of the same medium, and the shaker condition was 220 rpm, 30°C, 72 hours. After the cultivation was completed, the bacteria were collected by centrifugation at 12000 rpm. RNA extraction (Trizol method);
[0034] (2) Using RNA as a template to reverse transcribe and synthesize a cDNA sequence;
[0035] 2. Acquisition of deoxynivalenol degrading enzyme gene
[0036] Using the cDNA sequence obtained in step 1 as a template, a PCR reaction was performed under the guidance of primers 1 and 2 to a...
Embodiment 2
[0040] Embodiment 2: Cloning of mycotoxin ZEN degrading enzyme gene ZDase
[0041] PCR amplification was carried out using the standard strain G. pink ACCC No. 31535 genomic DNA as a template. Design the following primers to introduce EcoRI and NotI recognition sites (underlined) respectively
[0042] Upstream primer P3: 5'-CCG GAA TTC ATG CGC ACT CGC AGC ACAAT-3'
[0043] Downstream primer P4: 5'-ATAAGAAT GCG GCC G C AAGA TAC TTC TGC GTAAAT T-3'
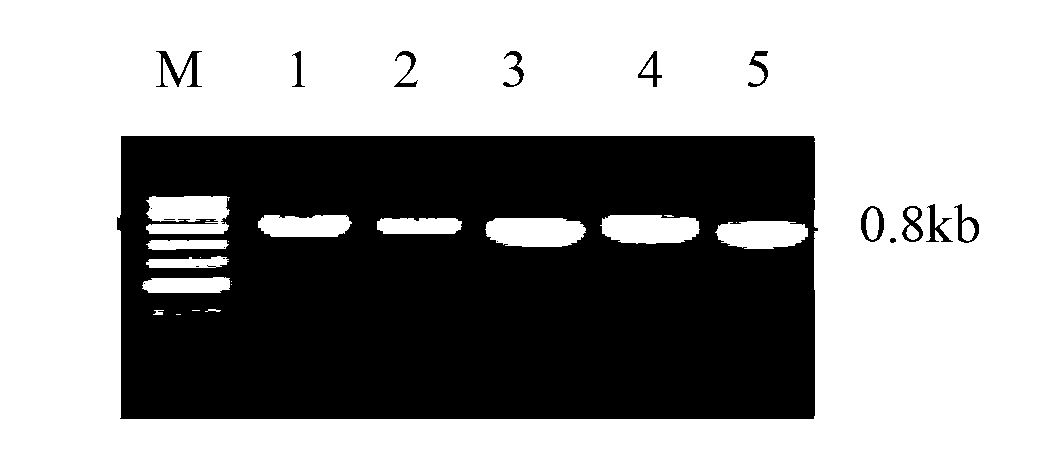
[0044] The PCR reaction contains 25 μL KOD enzyme Mix in 50 μL PCR reaction mixture, 2 μL each of 12.5 μmoL / L primers, 10 μL template, and 11 μL sterile water to adjust to 50 μL. Conditions are 94 ° C, denaturation 5 min, cycle parameters: 94 ° C denaturation 40 s; Anneal at 54°C for 1 min; extend at 68°C for 1 min; 30 cycles; extend at 68°C for 10 min to complete the extension of the product. The obtained PCR products were subjected to 1.0% agarose gel electrophoresis, and the electrophoresis results were as follows: figu...
Embodiment 3
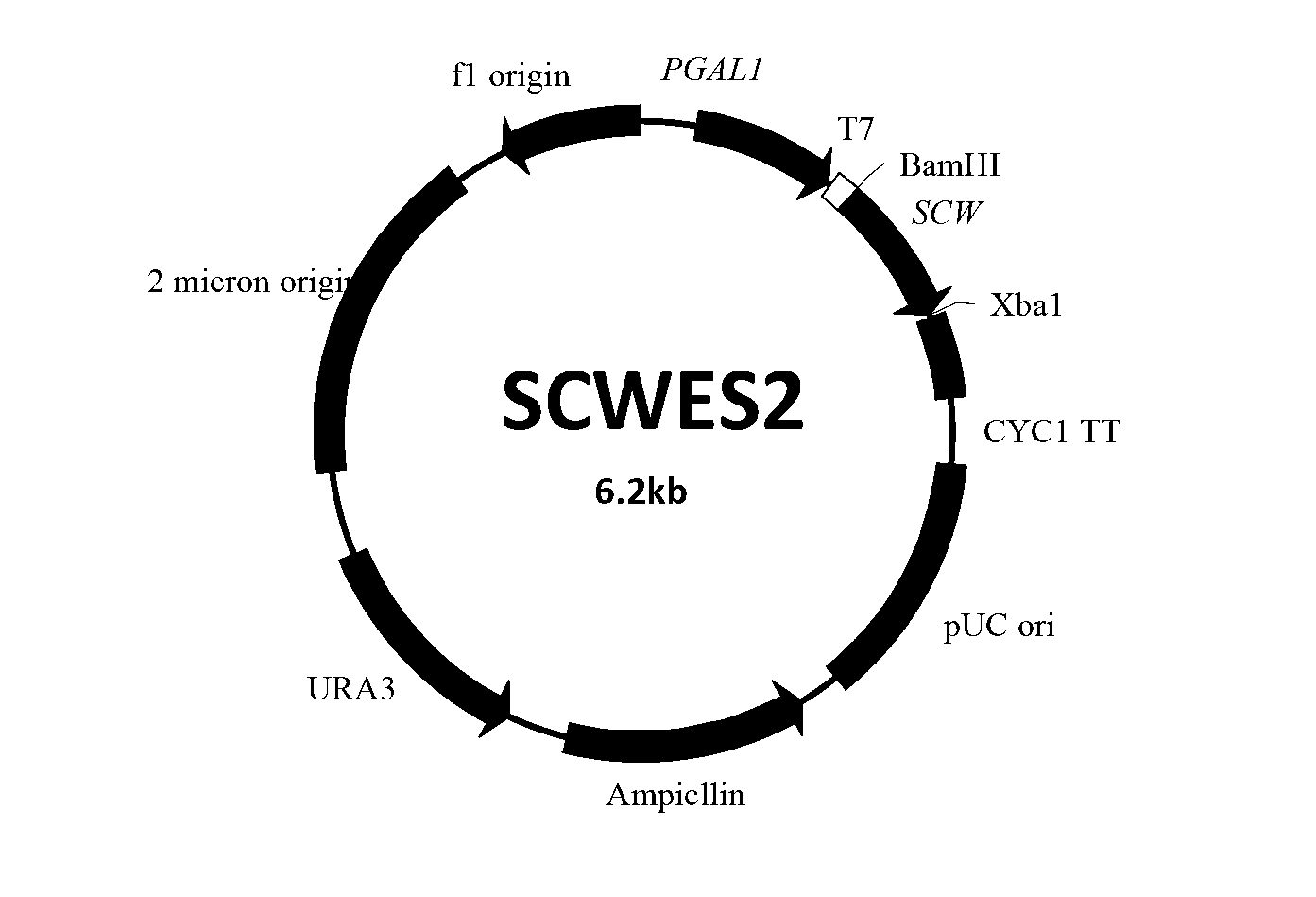
[0045] Example 3: Construction and verification of recombinant expression vectors
[0046] 1. Construction and verification of recombinant expression vector with exocrine ability
[0047] According to the reported sequence of the exocrine protein α mating factor coding gene on the exocrine expression vector of Pichia pastoris (GenBank Accession No. HQ398363.1 of the SCW gene sequence), the following primers were designed to introduce BamHI and XbaI recognition sites (underlined respectively) out)
[0048] Upstream primer P5: 5'-CGC GGATCC ATGAGATTTCCTTCAATTTT-3'
[0049] Downstream primer P6: 5'-GC TCTAGA TTAATTCGCGGCCGCCCTAG-3′.
[0050] The plasmid DNA of Pichia pastoris expression vector pPIC9k (purchased from Invitrogen) was used as a template for PCR amplification. For PCR reaction, 50 μL of PCR reaction mixture contains 25 μL of KOD enzyme Mix, 2 μL of 12.5 μmoL / L primers, 10 μL of template, and 11 μL of sterile water to adjust to 50 μL. Conditions are 94°C, denat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com