Method for establishing number of Fusarium sp. copies in rhizosphere soil in growth period of transgenic rice by fluorescence real-time quantitative PCR (polymerase chain reaction)
A transgenic wheat, real-time quantitative technology, applied in biochemical equipment and methods, fluorescence/phosphorescence, microbial measurement/inspection, etc., can solve problems that have not been reported, and achieve simple identification methods, high amplification efficiency, and good specificity sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Select the specific primer of Fusarium, and this primer sequence comes from Kamel A. Abd-Elsalam etc. (PCR identification of Fusarium genus based on nuclear ribosomal-DNA sequence data, African Journal of Biotechnology Vol. 2 (4), pp. 82-85 , April 2003)
[0028] SEQ ID No.2: ITS-Fu-F: 5'-CAACTCCCAAACCCCTGTGA-3';
[0029] SEQ ID No. 3: ITS-Fu-R: 5'-GCGACGATTACCAGTAACGA-3'.
[0030] In the transgenic wheat evaluation base of Jiangsu Academy of Agricultural Sciences, 64㎡ was selected as the experimental base, and 5 sampling points on the diagonal line were established in the field, and positioning marks were set during the experiment, and the soil samples collected at each point before sowing were blank. Control each sampling 0.5g at each point, sow the transgenic wheat line N12-1 in the field, and collect roots at the above-mentioned sampling soil points during the growth period of wheat at the seedling stage, turning green stage, jointing stage, filling stage, and matu...
Embodiment 2
[0038] Primer Specificity Detection
[0039] Select five kinds of pathogenic fungi commonly found in soil, Fusarium graminearum, Fusarium nivale, Fusarium axysporum, Rhizoctonia cerealis and Rhizoctonia solani Bacteria (Rhizoctonia solani) to verify the primers, the specific steps are:
[0040] (1) Extract the total DNA of the pure culture of these bacteria as a template
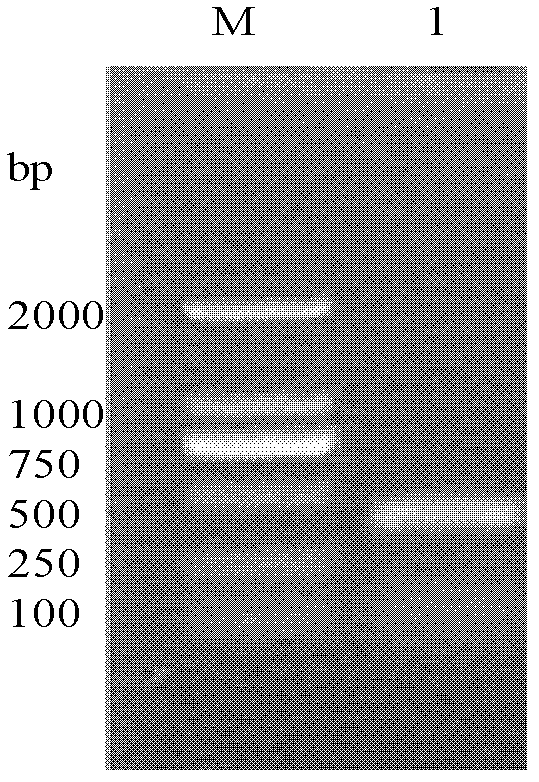
[0041] (2) Using the constructed plasmid DNA, the specific fragment with a size of about 400 bp obtained in Example 1 was recovered from agarose gel and connected to the pMD-19T vector (TaKaRa, Dalian) for cloning and transformation.
[0042] (3) Using wheat rhizosphere soil DNA as a positive control, Real-Time QPCR amplification was performed referring to the conditions of Example 1. The results showed that only in Fusarium graminearum, Fusarium nivale, and Fusarium oxysporum had DNA amplification products, and the other two bacteria had no corresponding amplification products. The results showed that th...
Embodiment 3
[0046] Establishment and detection analysis of fluorescent quantitative PCR system
[0047] Specific primer pair is identical with embodiment 1
[0048] (1) Preparation of Fusarium DNA template required for making standard curve
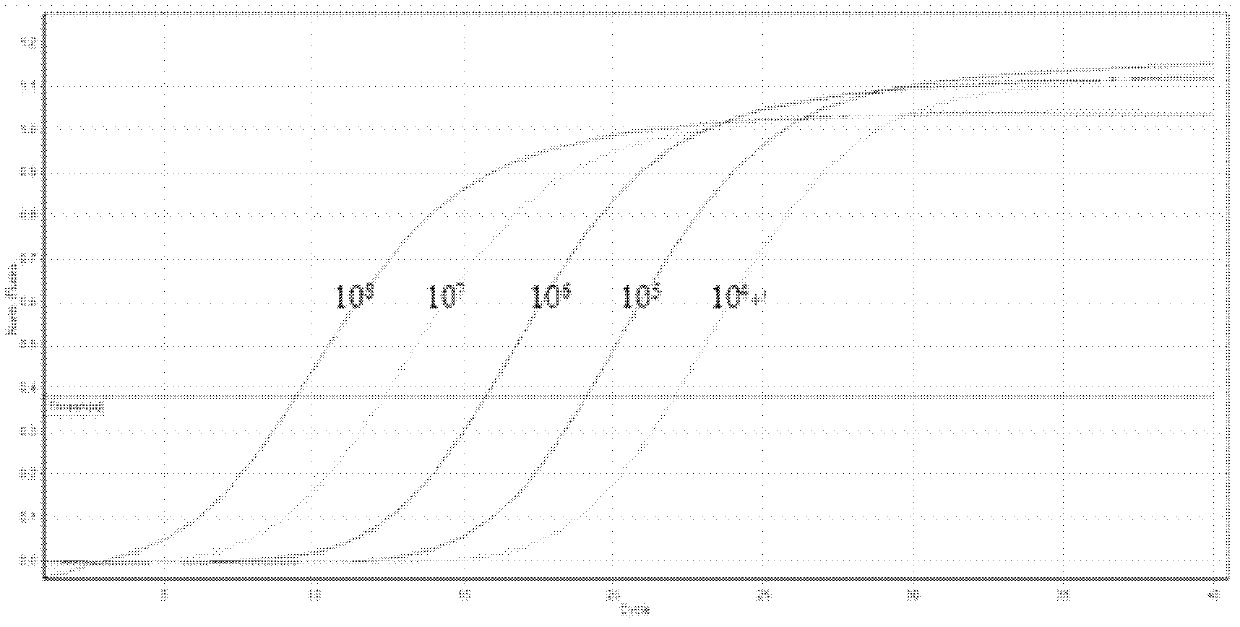
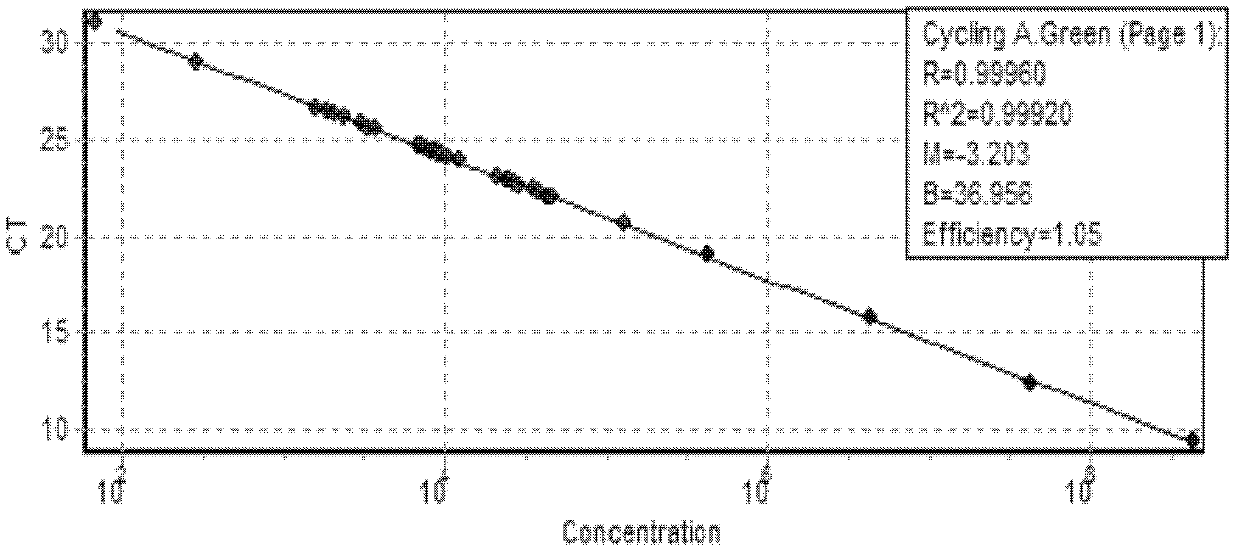
[0049] The product obtained in Example 1 was recovered by agarose gel and connected to the pMD-19T vector (TaKaRa, Dalian). After clone screening, sequencing analysis was performed. The sequencing result (SEQ ID No.1) showed that it was consistent with multiple Fusarium species in GenBank. The sequence is 100% homologous, indicating that the obtained sequence is correct, and the positive plasmid can be used to make plasmid standards. Use ultraviolet spectrophotometer to measure that this standard plasmid concentration is 4.30 * 10 10 copies / μl, the initial standard plasmid solution was serially diluted 10 times.
[0050] (2) Calculation of standard plasmid concentration
[0051] Extract the plasmids of the positive clones verified by sequencing,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com