Nucleic acid aptamer combining hepatitis B virus surface antigen and sequence thereof
A technology of hepatitis B virus and surface antigen, which is applied in the fields of genetic engineering and biomedicine, can solve the problems of chronic HBV infection and the lack of long-term effective antiviral drugs, and achieves the effect of simple method and low cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1H1
[0017] Example 1H1 protein specifically binds to the SELEX screening of RNA aptamers
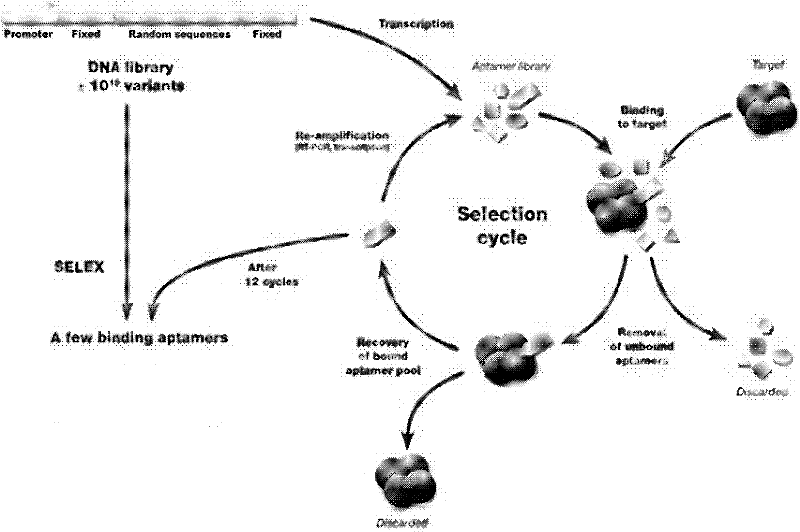
[0018] SELEX screening process such as figure 1 As shown, chemically synthesized initial oligonucleotide random library and primers, the sequence is as follows: 5'-TTAATACGACTCACTATAGTTGATTGCGTGTCAATCATGG-25N-GGTCATGTGTATGTTGGGGATTAGGACCTGATTGAGTTCAGCCCACATAC-3' (25N represents 25 random nucleotides);
[0019] Primer 1: 5′-TTAATACGACTCACTATAGTTGATTGCGTGTCAATC-3′,
[0020] Primer 2: 5'-GTATGTGGGCTGAACTCAAT-3'. The single-stranded DNA library is amplified into double-stranded DNA, and the product is subjected to 2% agarose gel electrophoresis and then gel-cut to recover and purify; the recovered double-stranded DNA is used as a template to transcribe a single-stranded RNA random library in vitro, and the transcript is purified by PAGE . 68 μg RNA library was reverse-screened with nitrocellulose membrane to remove membrane-bound RNA molecules, then incubated with 2 μg HBsAg at 37°C for 40 mi...
Embodiment 2
[0021] Secondary structure analysis of embodiment 2 RNA aptamers
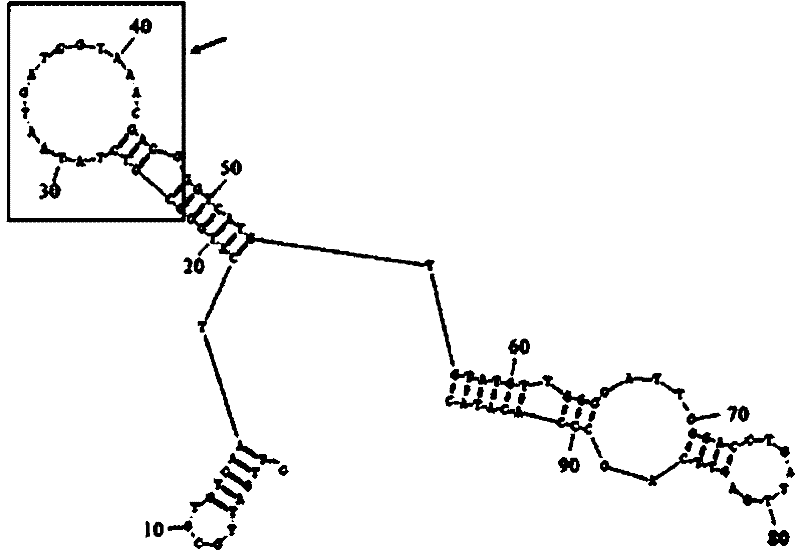
[0022] The library obtained in the 12th round of screening was cloned into the pMD19-T vector, transformed into Escherichia coli DH5α, and 48 clones were randomly selected and sequenced. Obtain the sequence information of the screened aptamers, predict the RNA secondary structure of all sequencing clones through the structure prediction software RNA Structure Program, and obtain an aptamer that can form a special stem-loop in the random sequence region (23-47nt) structure, such as figure 2 shown.
Embodiment 3
[0023] Example 3 Obtaining of HBsAg binding suitable gametes with high specificity and high affinity
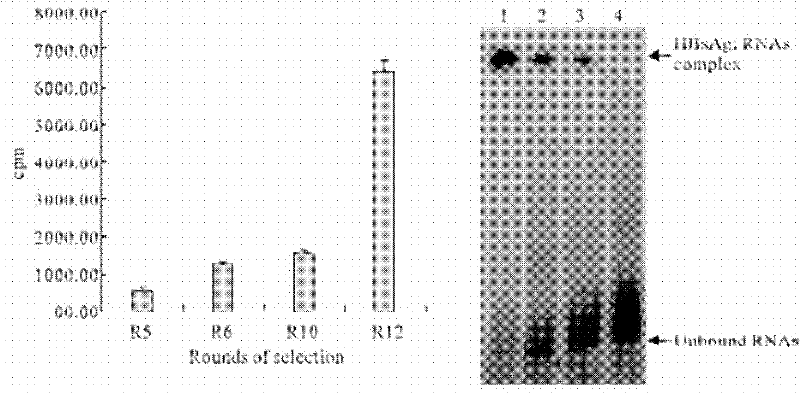
[0024] Take 1 μg of the RNA aptamers with the above secondary structures, digest them with calf intestinal alkaline phosphatase (CIP) at 37°C for 1 h, purify and recover the dephosphorylated RNA; label [γ- 32 P]ATP at the end of dephosphorylated RNA molecules. 10nmol of radioactively labeled RNA aptamers were incubated with different concentrations (10-200nM) of HBsAg at 37°C for 40min, and the reaction solution of each group was filtered through a nitrocellulose membrane, the filter membrane was washed, dried, and the filter membrane was measured by a liquid scintillation counter. The residual radioactive dose on the same sample was measured twice in parallel. The dissociation constants of each aptamer and HBsAg were calculated, and the aptamer with the highest affinity was obtained, which was named S-A22. 32 P-marked fifth-round library R5, sixth-round library R6, tenth-r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com